MARINE-EXPRESS: taking advantage of high throughput cloning and expression strategies for the post-genomic analysis of marine organisms
- PMID: 20546566
- PMCID: PMC2897777
- DOI: 10.1186/1475-2859-9-45
MARINE-EXPRESS: taking advantage of high throughput cloning and expression strategies for the post-genomic analysis of marine organisms
Abstract
Background: The production of stable and soluble proteins is one of the most important steps prior to structural and functional studies of biological importance. We investigated the parallel production in a medium throughput strategy of genes coding for proteins from various marine organisms, using protocols that involved recombinatorial cloning, protein expression screening and batch purification. This strategy was applied in order to respond to the need for post-genomic validation of the recent success of a large number of marine genomic projects. Indeed, the upcoming challenge is to go beyond the bioinformatic data, since the bias introduced through the genomes of the so called model organisms leads to numerous proteins of unknown function in the still unexplored world of the oceanic organisms.
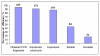
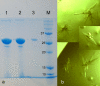
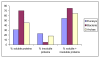
Results: We present here the results of expression tests for 192 targets using a 96-well plate format. Genes were PCR amplified and cloned in parallel into expression vectors pFO4 and pGEX-4T-1, in order to express proteins N-terminally fused to a six-histidine-tag and to a GST-tag, respectively. Small-scale expression and purification permitted isolation of 84 soluble proteins and 34 insoluble proteins, which could also be used in refolding assays. Selected examples of proteins expressed and purified to a larger scale are presented.
Conclusions: The objective of this program was to get around the bottlenecks of soluble, active protein expression and crystallization for post-genomic validation of a number of proteins that come from various marine organisms. Multiplying the constructions, vectors and targets treated in parallel is important for the success of a medium throughput strategy and considerably increases the chances to get rapid access to pure and soluble protein samples, needed for the subsequent biochemical characterizations. Our set up of a medium throughput strategy applied to genes from marine organisms had a mean success rate of 44% soluble protein expression from marine bacteria, archaea as well as eukaryotic organisms. This success rate compares favorably with other protein screening projects, particularly for eukaryotic proteins. Several purified targets have already formed the base for experiments aimed at post-genomic validation.
Figures



Similar articles
-
Pilot studies on the parallel production of soluble mouse proteins in a bacterial expression system.J Struct Funct Genomics. 2005;6(1):13-20. doi: 10.1007/s10969-005-0462-7. J Struct Funct Genomics. 2005. PMID: 15965734
-
Pichia pastoris is a valuable host for the expression of genes encoding membrane proteins from the hyperthermophilic Archeon Pyrococcus abyssi.Extremophiles. 2007 Mar;11(2):403-13. doi: 10.1007/s00792-006-0036-z. Epub 2006 Nov 8. Extremophiles. 2007. PMID: 17091222
-
Cloning, purification and crystallization of a Walker-type Pyrococcus abyssi ATPase family member.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 Oct 1;61(Pt 10):925-7. doi: 10.1107/S174430910502868X. Epub 2005 Sep 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005. PMID: 16511197 Free PMC article.
-
Tuning different expression parameters to achieve soluble recombinant proteins in E. coli: advantages of high-throughput screening.Biotechnol J. 2011 Jun;6(6):715-30. doi: 10.1002/biot.201100025. Epub 2011 May 12. Biotechnol J. 2011. PMID: 21567962 Review.
-
[Homologous protein domains in superkingdoms Archaea, Bacteria, and Eukaryota and the problem of the origin of eukaryotes].Izv Akad Nauk Ser Biol. 2005 Jul-Aug;(4):389-400. Izv Akad Nauk Ser Biol. 2005. PMID: 16212260 Review. Russian.
Cited by
-
Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate.J Biol Chem. 2013 Aug 9;288(32):23021-37. doi: 10.1074/jbc.M113.467217. Epub 2013 Jun 19. J Biol Chem. 2013. PMID: 23782694 Free PMC article.
-
Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans.J Biol Chem. 2012 Aug 31;287(36):30571-84. doi: 10.1074/jbc.M112.377184. Epub 2012 Jul 9. J Biol Chem. 2012. PMID: 22778272 Free PMC article.
-
Double blind microarray-based polysaccharide profiling enables parallel identification of uncharacterized polysaccharides and carbohydrate-binding proteins with unknown specificities.Sci Rep. 2018 Feb 6;8(1):2500. doi: 10.1038/s41598-018-20605-9. Sci Rep. 2018. PMID: 29410423 Free PMC article.
-
High throughput screening identifies disulfide isomerase DsbC as a very efficient partner for recombinant expression of small disulfide-rich proteins in E. coli.Microb Cell Fact. 2013 Apr 22;12:37. doi: 10.1186/1475-2859-12-37. Microb Cell Fact. 2013. PMID: 23607455 Free PMC article.
-
Regulation of alginate catabolism involves a GntR family repressor in the marine flavobacterium Zobellia galactanivorans DsijT.Nucleic Acids Res. 2020 Aug 20;48(14):7786-7800. doi: 10.1093/nar/gkaa533. Nucleic Acids Res. 2020. PMID: 32585009 Free PMC article.
References
-
- Cohen G, Barbe V, Flament D, Galperin M, Heilig R, Lecompte O, Poch O, Prieur D, Quérellou J, Ripp R, Thierry JC, Van der Oost J, Weissenbach J, Zivanovic Y, Forterre P. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol Microbiol. 2003;47:1495–1512. doi: 10.1046/j.1365-2958.2003.03381.x. - DOI - PubMed
-
- Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W, Gade D, Beck A, Borzym K, Heitmann K, Rabus R, Schlesner H, Amann R, Reinhardt R. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA. 2003;100:8298–8303. doi: 10.1073/pnas.1431443100. - DOI - PMC - PubMed
-
- Peters AF, Marie D, Scornet D, Kloareg B, Cock JM. Proposal of Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) as a model organism for brown algal genetics and genomics. J Phycol. 2004;40:1079–1088. doi: 10.1111/j.1529-8817.2004.04058.x. - DOI
-
- Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, Saeys Y, Wuyts J, Jabbari K, Bowler C, Panaud O, Piegu B, Ball SG, Ral JP, Bouget FY, Piganeau G, De Baets B, Picard A, Delseny M, Demaille J, Van de Peer Y, Moreau H. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. - DOI - PMC - PubMed
-
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

