Genomic analysis of carboxyl/cholinesterase genes in the silkworm Bombyx mori
- PMID: 20546589
- PMCID: PMC3017765
- DOI: 10.1186/1471-2164-11-377
Genomic analysis of carboxyl/cholinesterase genes in the silkworm Bombyx mori
Abstract
Background: Carboxyl/cholinesterases (CCEs) have pivotal roles in dietary detoxification, pheromone or hormone degradation and neurodevelopment. The recent completion of genome projects in various insect species has led to the identification of multiple CCEs with unknown functions. Here, we analyzed the phylogeny, expression and genomic distribution of 69 putative CCEs in the silkworm, Bombyx mori (Lepidoptera: Bombycidae).
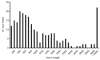
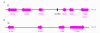
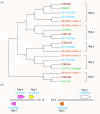
Results: A phylogenetic tree of CCEs in B. mori and other lepidopteran species was constructed. The expression pattern of each B. mori CCE was also investigated by a search of an expressed sequence tag (EST) database, and the relationship between phylogeny and expression was analyzed. A large number of B. mori CCEs were identified from a midgut EST library. CCEs expressed in the midgut formed a cluster in the phylogenetic tree that included not only B. mori genes but also those of other lepidopteran species. The silkworm, and possibly also other lepidopteran species, has a large number of CCEs, and this might be a consequence of the large cluster of midgut CCEs. Investigation of intron-exon organization in B. mori CCEs revealed that their positions and splicing site phases were strongly conserved. Several B. mori CCEs, including juvenile hormone esterase, not only showed clustering in the phylogenetic tree but were also closely located on silkworm chromosomes. We investigated the phylogeny and microsynteny of neuroligins in detail, among many CCEs. Interestingly, we found the evolution of this gene appeared not to be conserved between B. mori and other insect orders.
Conclusions: We analyzed 69 putative CCEs from B. mori. Comparison of these CCEs with other lepidopteran CCEs indicated that they had conserved expression and function in this insect order. The analyses showed that CCEs were unevenly distributed across the genome of B. mori and suggested that neuroligins may have a distinct evolutionary history from other insect order. It is possible that such an uneven genomic distribution and a unique neuroligin evolution are shared with other lepidopteran insects. Our genomic analysis has provided novel information on the CCEs of the silkworm, which will be of value to understanding the biology, physiology and evolution of insect CCEs.
Figures



References
-
- Oakeshott JG, Claudianos C, Campbell PM, Newcomb RD, Russell RJ. In: Comprehensive Molecular Insect Science-Pharmacology. Gilbert LI, Iatrou K, Gill SS, editor. Vol. 5. Oxford: Elsevier; 2005. Biochemical genetics and genomics of insect esterases; pp. 309–381. full_text.
-
- Riddiford LM. Cellular and molecular actions of juvenile hormone 1. General considerations and premetamorphic actions. Advances in Insect Physiology. 1994;24:213–274. full_text.
-
- Wyatt GR, Davey KG. Cellular and molecular actions of juvenile hormone 2. Roles of juvenile hormone in adult insects. Advances in Insect Physiology. 1996;26:1–155. full_text.
-
- Jallon JM, Wicker-Thomas C. In: Insect Pheromone Biochemistry and Molecular Biology. Blomquist GJ, Vogt RG, editor. Oxford: Elsevier; 2003. Genetic studies on pheromone production in Drosophila; pp. 253–281. full_text.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

