A gene network switch enhances the oxidative capacity of ovine skeletal muscle during late fetal development
- PMID: 20546621
- PMCID: PMC2894804
- DOI: 10.1186/1471-2164-11-378
A gene network switch enhances the oxidative capacity of ovine skeletal muscle during late fetal development
Abstract
Background: The developmental transition between the late fetus and a newborn animal is associated with profound changes in skeletal muscle function as it adapts to the new physiological demands of locomotion and postural support against gravity. The mechanisms underpinning this adaption process are unclear but are likely to be initiated by changes in hormone levels. We tested the hypothesis that this developmental transition is associated with large coordinated changes in the transcription of skeletal muscle genes.
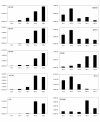
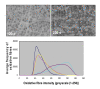
Results: Using an ovine model, transcriptional profiling was performed on Longissimus dorsi skeletal muscle taken at three fetal developmental time points (80, 100 and 120 d of fetal development) and two postnatal time points, one approximately 3 days postpartum and a second at 3 months of age. The developmental time course was dominated by large changes in expression of 2,471 genes during the interval between late fetal development (120 d fetal development) and 1-3 days postpartum. Analysis of the functions of genes that were uniquely up-regulated in this interval showed strong enrichment for oxidative metabolism and the tricarboxylic acid cycle indicating enhanced mitochondrial activity. Histological examination of tissues from these developmental time points directly confirmed a marked increase in mitochondrial activity between the late fetal and early postnatal samples. The promoters of genes that were up-regulated during this fetal to neonatal transition were enriched for estrogen receptor 1 and estrogen related receptor alpha cis-regulatory motifs. The genes down-regulated during this interval highlighted de-emphasis of an array of functions including Wnt signaling, cell adhesion and differentiation. There were also changes in gene expression prior to this late fetal--postnatal transition and between the two postnatal time points. The former genes were enriched for functions involving the extracellular matrix and immune response while the latter principally involved functions associated with transcriptional regulation of metabolic processes.
Conclusions: It is concluded that during late skeletal muscle development there are substantial and coordinated changes in the transcription of a large number of genes many of which are probably triggered by increased estrogen levels. These changes probably underpin the adaption of muscle to new physiological demands in the postnatal environment.
Figures
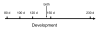


Similar articles
-
Genomic architecture of histone 3 lysine 27 trimethylation during late ovine skeletal muscle development.Anim Genet. 2014 Jun;45(3):427-38. doi: 10.1111/age.12145. Epub 2014 Mar 27. Anim Genet. 2014. PMID: 24673416 Free PMC article.
-
Histological and transcriptome-wide level characteristics of fetal myofiber hyperplasia during the second half of gestation in Texel and Ujumqin sheep.BMC Genomics. 2011 Aug 14;12:411. doi: 10.1186/1471-2164-12-411. BMC Genomics. 2011. PMID: 21838923 Free PMC article.
-
Identification of a gene network contributing to hypertrophy in callipyge skeletal muscle.Physiol Genomics. 2007 Feb 12;28(3):253-72. doi: 10.1152/physiolgenomics.00121.2006. Epub 2006 Oct 31. Physiol Genomics. 2007. PMID: 17077277
-
Co-expression analysis of fetal weight-related genes in ovine skeletal muscle during mid and late fetal development stages.Int J Biol Sci. 2014 Sep 11;10(9):1039-50. doi: 10.7150/ijbs.9737. eCollection 2014. Int J Biol Sci. 2014. PMID: 25285036 Free PMC article.
-
Gene expression studies of developing bovine longissimus muscle from two different beef cattle breeds.BMC Dev Biol. 2007 Aug 16;7:95. doi: 10.1186/1471-213X-7-95. BMC Dev Biol. 2007. PMID: 17697390 Free PMC article.
Cited by
-
Brown to White Fat Transition Overlap With Skeletal Muscle During Development of Larger Mammals: Is it a Coincidence?J Endocr Soc. 2022 Sep 27;6(12):bvac151. doi: 10.1210/jendso/bvac151. eCollection 2022 Oct 26. J Endocr Soc. 2022. PMID: 36325536 Free PMC article. Review.
-
Genomic architecture of histone 3 lysine 27 trimethylation during late ovine skeletal muscle development.Anim Genet. 2014 Jun;45(3):427-38. doi: 10.1111/age.12145. Epub 2014 Mar 27. Anim Genet. 2014. PMID: 24673416 Free PMC article.
-
Muscle transcriptome provides the first insight into the dynamics of gene expression with progression of age in sheep.Sci Rep. 2021 Nov 16;11(1):22360. doi: 10.1038/s41598-021-01848-5. Sci Rep. 2021. PMID: 34785720 Free PMC article.
-
Maternal and paternal genomes differentially affect myofibre characteristics and muscle weights of bovine fetuses at midgestation.PLoS One. 2013;8(1):e53402. doi: 10.1371/journal.pone.0053402. Epub 2013 Jan 14. PLoS One. 2013. PMID: 23341941 Free PMC article.
-
Protein composition of the muscle mitochondrial reticulum during postnatal development.J Physiol. 2019 May;597(10):2707-2727. doi: 10.1113/JP277579. Epub 2019 Apr 8. J Physiol. 2019. PMID: 30919448 Free PMC article.
References
-
- Maier A, McEwan JC, Dodds KG, Fischman DA, Fitzsimons RB, Harris AJ. Myosin heavy chain composition of single fibres and their origins and distribution in developing fascicles of sheep tibialis cranialis muscles. Journal of muscle research and cell motility. 1992;13(5):551–572. doi: 10.1007/BF01737997. - DOI - PubMed
-
- Greenwood PL, Gardner GE, Hegarty RS. Lamb myofibre characteristics are influenced by sire estimated breeding values and pastoral nutritional system. Australian Journal of Agricultural Research. 2006;57:627–639. doi: 10.1071/AR04318. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

