Pripper: prediction of caspase cleavage sites from whole proteomes
- PMID: 20546630
- PMCID: PMC2893604
- DOI: 10.1186/1471-2105-11-320
Pripper: prediction of caspase cleavage sites from whole proteomes
Abstract
Background: Caspases are a family of proteases that have central functions in programmed cell death (apoptosis) and inflammation. Caspases mediate their effects through aspartate-specific cleavage of their target proteins, and at present almost 400 caspase substrates are known. There are several methods developed to predict caspase cleavage sites from individual proteins, but currently none of them can be used to predict caspase cleavage sites from multiple proteins or entire proteomes, or to use several classifiers in combination. The possibility to create a database from predicted caspase cleavage products for the whole genome could significantly aid in identifying novel caspase targets from tandem mass spectrometry based proteomic experiments.
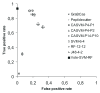
Results: Three different pattern recognition classifiers were developed for predicting caspase cleavage sites from protein sequences. Evaluation of the classifiers with quality measures indicated that all of the three classifiers performed well in predicting caspase cleavage sites, and when combining different classifiers the accuracy increased further. A new tool, Pripper, was developed to utilize the classifiers and predict the caspase cut sites from an arbitrary number of input sequences. A database was constructed with the developed tool, and it was used to identify caspase target proteins from tandem mass spectrometry data from two different proteomic experiments. Both known caspase cleavage products as well as novel cleavage products were identified using the database demonstrating the usefulness of the tool. Pripper is not restricted to predicting only caspase cut sites, but it gives the possibility to scan protein sequences for any given motif(s) and predict cut sites once a suitable cut site prediction model for any other protease has been developed. Pripper is freely available and can be downloaded from http://users.utu.fi/mijopi/Pripper.
Conclusions: We have developed Pripper, a tool for reading an arbitrary number of proteins in FASTA format, predicting their caspase cleavage sites and outputting the cleaved sequences to a new FASTA format sequence file. We show that Pripper is a valuable tool in identifying novel caspase target proteins from modern proteomics experiments.
Figures


Similar articles
-
Cascleave: towards more accurate prediction of caspase substrate cleavage sites.Bioinformatics. 2010 Mar 15;26(6):752-60. doi: 10.1093/bioinformatics/btq043. Epub 2010 Feb 3. Bioinformatics. 2010. PMID: 20130033
-
A multi-factor model for caspase degradome prediction.BMC Genomics. 2009 Dec 3;10 Suppl 3(Suppl 3):S6. doi: 10.1186/1471-2164-10-S3-S6. BMC Genomics. 2009. PMID: 19958504 Free PMC article.
-
Cascleave 2.0, a new approach for predicting caspase and granzyme cleavage targets.Bioinformatics. 2014 Jan 1;30(1):71-80. doi: 10.1093/bioinformatics/btt603. Epub 2013 Oct 21. Bioinformatics. 2014. PMID: 24149049
-
Toward computer-based cleavage site prediction of cysteine endopeptidases.Biol Chem. 2003 Jun;384(6):899-909. doi: 10.1515/BC.2003.101. Biol Chem. 2003. PMID: 12887057 Review.
-
The CASBAH: a searchable database of caspase substrates.Cell Death Differ. 2007 Apr;14(4):641-50. doi: 10.1038/sj.cdd.4402103. Epub 2007 Feb 2. Cell Death Differ. 2007. PMID: 17273173 Review.
Cited by
-
Prediction and Design of Protease Enzyme Specificity Using a Structure-Aware Graph Convolutional Network.bioRxiv [Preprint]. 2023 Feb 16:2023.02.16.528728. doi: 10.1101/2023.02.16.528728. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Sep 26;120(39):e2303590120. doi: 10.1073/pnas.2303590120. PMID: 36824945 Free PMC article. Updated. Preprint.
-
Knowledge-transfer learning for prediction of matrix metalloprotease substrate-cleavage sites.Sci Rep. 2017 Jul 18;7(1):5755. doi: 10.1038/s41598-017-06219-7. Sci Rep. 2017. PMID: 28720874 Free PMC article.
-
ReCGBM: a gradient boosting-based method for predicting human dicer cleavage sites.BMC Bioinformatics. 2021 Feb 10;22(1):63. doi: 10.1186/s12859-021-03993-0. BMC Bioinformatics. 2021. PMID: 33568063 Free PMC article.
-
Developing a powerful in silico tool for the discovery of novel caspase-3 substrates: a preliminary screening of the human proteome.BMC Bioinformatics. 2012 Jan 23;13:14. doi: 10.1186/1471-2105-13-14. BMC Bioinformatics. 2012. PMID: 22269041 Free PMC article.
-
Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning.Cell Host Microbe. 2023 Aug 9;31(8):1260-1274.e6. doi: 10.1016/j.chom.2023.07.001. Epub 2023 Jul 28. Cell Host Microbe. 2023. PMID: 37516110 Free PMC article.
References
-
- Song J, Tan H, Shen H, Mahmood K, Boyd SE, Webb GI, Akutsu T, Whisstock JC. Cascleave: towards more accurate prediction of caspase substrate cleavage sites. Bioinformatics. 2010. btq043. - PubMed
-
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

