Development of a dual-aptamer-based multiplex protein biosensor
- PMID: 20547050
- PMCID: PMC2891049
- DOI: 10.1016/j.bios.2010.04.034
Development of a dual-aptamer-based multiplex protein biosensor
Abstract
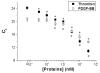
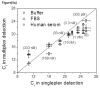
Parallel biosensors for proteins are becoming more essential for the thorough and systematic investigation of complex biological processes. These tools also enable improved clinical diagnoses relative to single-protein analyses due to their greater information content. If implemented correctly, affinity-based techniques can provide unique advantages in terms of sensitivity and flexibility. Aptamers are increasingly being used as the affinity reagents of choice for protein biosensing applications. Here, we describe the development and characterization of an aptamer-based method for parallel protein analyses that relies on recognition of the target protein by two unique aptamers targeting different epitopes on the protein. Our results show that the technique achieved simultaneous and quantitative detection of thrombin and platelet-derived growth factor-BB (PDGF-BB) with high specificity both in buffered solutions and in serum samples.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Lateral Flow Aptasensor for Simultaneous Detection of Platelet-Derived Growth Factor-BB (PDGF-BB) and Thrombin.Molecules. 2019 Feb 20;24(4):756. doi: 10.3390/molecules24040756. Molecules. 2019. PMID: 30791526 Free PMC article.
-
Thrombin-linked aptamer assay for detection of platelet derived growth factor BB on magnetic beads in a sandwich format.Talanta. 2016 Sep 1;158:159-164. doi: 10.1016/j.talanta.2016.05.037. Epub 2016 May 14. Talanta. 2016. PMID: 27343590
-
A SERS-LFA biosensor combined with aptamer recognition for simultaneous detection of thrombin and PDGF-BB in prostate cancer plasma.Nanotechnology. 2021 Aug 13;32(44). doi: 10.1088/1361-6528/ac1754. Nanotechnology. 2021. PMID: 34298537
-
Recent advances on aptamer-based biosensors to detection of platelet-derived growth factor.Biosens Bioelectron. 2018 Aug 15;113:58-71. doi: 10.1016/j.bios.2018.04.048. Epub 2018 Apr 22. Biosens Bioelectron. 2018. PMID: 29729560 Review.
-
Nucleic acid aptamers for biosensors and bio-analytical applications.Analyst. 2009 Sep;134(9):1765-75. doi: 10.1039/b905609m. Epub 2009 Jun 23. Analyst. 2009. PMID: 19684896 Review.
Cited by
-
Identification and Affinity Determination of Protein-Antibody and Protein-Aptamer Epitopes by Biosensor-Mass Spectrometry Combination.Int J Mol Sci. 2021 Nov 27;22(23):12832. doi: 10.3390/ijms222312832. Int J Mol Sci. 2021. PMID: 34884636 Free PMC article. Review.
-
Inhibition of Colorectal Cancer Cell Proliferation by Regulating Platelet-Derived Growth Factor B Signaling with a DNA Aptamer.Asian Pac J Cancer Prev. 2019 Feb 26;20(2):487-494. doi: 10.31557/APJCP.2019.20.2.487. Asian Pac J Cancer Prev. 2019. PMID: 30803211 Free PMC article.
-
Aptamer-functionalized microgel particles for protein detection.Anal Chem. 2011 Dec 1;83(23):9138-45. doi: 10.1021/ac202335u. Epub 2011 Nov 7. Anal Chem. 2011. PMID: 22017663 Free PMC article.
-
Streamlined circular proximity ligation assay provides high stringency and compatibility with low-affinity antibodies.Proc Natl Acad Sci U S A. 2018 Jan 30;115(5):E925-E933. doi: 10.1073/pnas.1718283115. Epub 2018 Jan 16. Proc Natl Acad Sci U S A. 2018. PMID: 29339495 Free PMC article.
-
Lateral Flow Aptasensor for Simultaneous Detection of Platelet-Derived Growth Factor-BB (PDGF-BB) and Thrombin.Molecules. 2019 Feb 20;24(4):756. doi: 10.3390/molecules24040756. Molecules. 2019. PMID: 30791526 Free PMC article.
References
-
- Bang GS, Cho S, Kim BG. Biosensors & bioelectronics. 2005;21(6):863–870. - PubMed
-
- Bock C, Coleman M, Collins B, Davis J, Foulds G, Gold L, Greef C, Heil J, Heilig JS, Hicke B, Hurst MN, Husar GM, Miller D, Ostroff R, Petach H, Schneider D, Vant-Hull B, Waugh S, Weiss A, Wilcox SK, Zichi D. Proteomics. 2004;4(3):609–618. - PubMed
-
- Breaker RR. Curr Opin Biotechnol. 2002;13(1):31–39. - PubMed
-
- Cho EJ, Collett JR, Szafranska AE, Ellington AD. Anal Chim Acta. 2006;564(1):82–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

