Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract
- PMID: 20547745
- PMCID: PMC2937465
- DOI: 10.1128/IAI.00386-10
Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract
Abstract
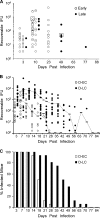
Chlamydia trachomatis is a human pathogen of global importance. An obstacle to studying the pathophysiology of human chlamydial disease is the lack of a suitable murine model of C. trachomatis infection. Mice are less susceptible to infection with human isolates due in part to innate mouse-specific host defense mechanisms to which human strains are sensitive. Another possible factor that influences the susceptibility of mice to infection is that human isolates are commonly cultivated in vitro prior to infection of mice; therefore, virulence genes could be lost as a consequence of negative selective pressure. We tested this hypothesis by infecting innate immunity-deficient C3H/HeJ female mice intravaginally with a human serovar D urogenital isolate that had undergone multiple in vitro passages. We observed early and late infection clearance phenotypes. Strains of each phenotype were isolated and then used to reinfect naïve mice. Following infection, the late-clearance strain was significantly more virulent. It caused unvarying infections of much longer durations with greater infectious burdens that naturally ascended to the upper genital tract, causing salpingitis. Despite contrasting in vivo virulence characteristics, the strains exhibited no differences in the results of in vitro infectivity assays or sensitivities to gamma interferon. Genome sequencing of the strains revealed mutations that localized to a single gene (CT135), implicating it as a critical virulence factor. Mutations in CT135 were not unique to serovar D but were also found in multiple oculogenital reference strains. Our findings provide new information about the pathogenomics of chlamydial infection and insights for improving murine models of infection using human strains.
Figures
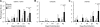




References
-
- Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111:1757-1769. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

