E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates
- PMID: 20547746
- PMCID: PMC2937443
- DOI: 10.1128/IAI.00344-10
E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates
Abstract
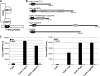
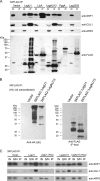
The intracellular bacterial pathogen Legionella pneumophila modulates a number of host processes during intracellular growth, including the eukaryotic ubiquitination machinery, which dictates the stability, activity, and/or localization of a large number of proteins. A number of L. pneumophila proteins contain eukaryotic-like motifs typically associated with ubiquitination. Central among these is a family of five F-box-domain-containing proteins of Legionella pneumophila. Each of these five proteins is translocated to the host cytosol by the Dot/Icm type IV protein translocation system during infection. We show that three of these proteins, LegU1, LegAU13, and LicA, interact with components of the host ubiquitination machinery in vivo. In addition, LegU1 and LegAU13 are integrated into functional Skp-Cullin-F-box (SCF) complexes that confer E3 ubiquitin ligase activity. LegU1 specifically interacts with and can direct the ubiquitination of the host chaperone protein BAT3. In a screen for additional L. pneumophila proteins that associate with LegU1 in mammalian cells, we identified the bacterial protein Lpg2160. We demonstrate that Lpg2160 also associates with BAT3 independently of LegU1. We show that Lpg2160 is a translocated substrate of the Dot/Icm system and contains a C-terminal translocation signal. We propose a model in which LegU1 and Lpg2160 may function redundantly or in concert to modulate BAT3 activity during the course of infection.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

