Distribution and phylogeny of immunoglobulin-binding protein G in Shiga toxin-producing Escherichia coli and its association with adherence phenotypes
- PMID: 20547747
- PMCID: PMC2916290
- DOI: 10.1128/IAI.00006-10
Distribution and phylogeny of immunoglobulin-binding protein G in Shiga toxin-producing Escherichia coli and its association with adherence phenotypes
Abstract
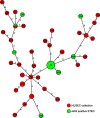
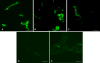
eibG in Shiga toxin-producing Escherichia coli (STEC) O91 encodes a protein (EibG) which binds human immunoglobulins G and A and contributes to bacterial chain-like adherence to human epithelial cells. We investigated the prevalence of eibG among STEC, the phylogeny of eibG, and eibG allelic variations and their impact on the adherence phenotype. eibG was found in 15.0% of 240 eae-negative STEC strains but in none of 157 eae-positive STEC strains. The 36 eibG-positive STEC strains belonged to 14 serotypes and to eight multilocus sequence types (STs), with serotype O91:H14/H(-) and ST33 being the most common. Sequences of the complete eibG gene (1,527 bp in size) from eibG-positive STEC resulted in 21 different alleles with 88.11% to 100% identity to the previously reported eibG sequence; they clustered into three eibG subtypes (eibG-alpha, eibG-beta, and eibG-gamma). Strains expressing EibG-alpha and EibG-beta displayed a mostly typical chain-like adherence pattern (CLAP), with formation of long chains on both human and bovine intestinal epithelial cells, whereas strains with EibG-gamma adhered in short chains, a pattern we termed atypical CLAP. The same adherence phenotypes were displayed by E. coli BL21(DE3) clones containing the respective eibG-alpha, eibG-beta, and eibG-gamma subtypes. We propose two possible evolutionary scenarios for eibG in STEC: a clonal development of eibG in strains with the same phylogenetic background or horizontal transfer of eibG between phylogenetically unrelated STEC strains.
Figures



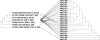



References
-
- Aldick, T., M. Bielaszewska, W. Zhang, J. Brockmeyer, H. Schmidt, A. W. Friedrich, K. S. Kim, M. A. Schmidt, and H. Karch. 2007. Hemolysin from Shiga toxin-negative Escherichia coli O26 strains injures microvascular endothelium. Microbes Infect. 9:282-290. - PubMed
-
- Bettelheim, K. A. 2007. The non-O157 Shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33:67-87. - PubMed
-
- Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

