Vitamin A metabolite, all-trans-retinoic acid, mediates alternative splicing of protein kinase C deltaVIII (PKCdeltaVIII) isoform via splicing factor SC35
- PMID: 20547768
- PMCID: PMC2923993
- DOI: 10.1074/jbc.M110.100735
Vitamin A metabolite, all-trans-retinoic acid, mediates alternative splicing of protein kinase C deltaVIII (PKCdeltaVIII) isoform via splicing factor SC35
Abstract
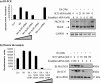
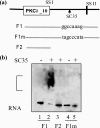
Vitamin A metabolite, all-trans-retinoic acid (RA), induces cell growth, differentiation, and apoptosis and has an emerging role in gene regulation and alternative splicing events. Protein kinase Cdelta (PKCdelta), a serine/threonine kinase, has a role in cell proliferation, differentiation, and apoptosis. We reported an alternatively spliced variant of human PKCdelta, PKCdeltaVIII that functions as a pro-survival protein (1). RA regulates the splicing and expression of PKCdeltaVIII via utilization of a downstream 5' splice site of exon 10 on PKCdelta pre-mRNA. Here, we further elucidate the molecular mechanisms involved in RA regulation of alternative splicing of PKCdeltaVIII mRNA. Overexpression and knockdown of the splicing factor SC35 (i.e. SRp30b) indicated that it is involved in PKCdeltaVIII alternative splicing. To identify the cis-elements involved in 5' splice site selection we cloned a minigene, which included PKCdelta exon 10 and its flanking introns in the pSPL3 splicing vector. Alternative 5' splice site utilization in the minigene was promoted by RA. Further, co-transfection of SC35 with PKCdelta minigene promoted selection of 5' splice site II. Mutation of the SC35 binding site in the PKCdelta minigene abolished RA-mediated utilization of 5' splice splice II. RNA binding assays demonstrated that the enhancer element downstream of PKCdelta exon 10 is a SC35 cis-element. We conclude that SC35 is pivotal in RA-mediated PKCdelta pre-mRNA alternative splicing. This study demonstrates how a nutrient, vitamin A, via its metabolite RA, regulates alternative splicing and thereby gene expression of the pro-survival protein PKCdeltaVIII.
Figures

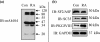




Similar articles
-
Identification of a novel antiapoptotic human protein kinase C delta isoform, PKCdeltaVIII in NT2 cells.Biochemistry. 2008 Jan 15;47(2):787-97. doi: 10.1021/bi7019782. Epub 2007 Dec 20. Biochemistry. 2008. PMID: 18092819
-
Role of alternatively spliced, pro-survival Protein Kinase C delta VIII (PKCδVIII) in ovarian cancer.FASEB Bioadv. 2021 Dec 10;4(4):235-253. doi: 10.1096/fba.2021-00090. eCollection 2022 Apr. FASEB Bioadv. 2021. PMID: 35415459 Free PMC article.
-
SC35 promotes splicing of the C5-V6-C6 isoform of CD44 pre-mRNA.Oncol Rep. 2014 Jan;31(1):273-9. doi: 10.3892/or.2013.2812. Epub 2013 Oct 24. Oncol Rep. 2014. PMID: 24173428 Free PMC article.
-
The mitochondrial PKCδ/retinol signal complex exerts real-time control on energy homeostasis.Biochim Biophys Acta Mol Cell Biol Lipids. 2020 Nov;1865(11):158614. doi: 10.1016/j.bbalip.2020.158614. Epub 2020 Jan 10. Biochim Biophys Acta Mol Cell Biol Lipids. 2020. PMID: 31927141 Free PMC article. Review.
-
Alternative splicing and cell survival: from tissue homeostasis to disease.Cell Death Differ. 2016 Dec;23(12):1919-1929. doi: 10.1038/cdd.2016.91. Epub 2016 Sep 30. Cell Death Differ. 2016. PMID: 27689872 Free PMC article. Review.
Cited by
-
Role and Mechanism of PKC-δ for Cardiovascular Disease: Current Status and Perspective.Front Cardiovasc Med. 2022 Feb 15;9:816369. doi: 10.3389/fcvm.2022.816369. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35242825 Free PMC article. Review.
-
SEL1L SNP rs12435998, a predictor of glioblastoma survival and response to radio-chemotherapy.Oncotarget. 2015 May 20;6(14):12452-67. doi: 10.18632/oncotarget.3611. Oncotarget. 2015. PMID: 25948789 Free PMC article.
-
MALAT1 in Human Adipose Stem Cells Modulates Survival and Alternative Splicing of PKCδII in HT22 Cells.Endocrinology. 2017 Jan 1;158(1):183-195. doi: 10.1210/en.2016-1819. Endocrinology. 2017. PMID: 27841943 Free PMC article.
-
Heat stress affects tassel development and reduces the kernel number of summer maize.Front Plant Sci. 2023 Jun 7;14:1186921. doi: 10.3389/fpls.2023.1186921. eCollection 2023. Front Plant Sci. 2023. PMID: 37351221 Free PMC article.
-
Protein kinase C δ (PKCδ) splice variants modulate apoptosis pathway in 3T3L1 cells during adipogenesis: identification of PKCδII inhibitor.J Biol Chem. 2013 Sep 13;288(37):26834-46. doi: 10.1074/jbc.M113.482638. Epub 2013 Jul 31. J Biol Chem. 2013. PMID: 23902767 Free PMC article.
References
-
- Jiang K., Apostolatos A. H., Ghansah T., Watson J. E., Vickers T., Cooper D. R., Epling-Burnette P. K., Patel N. A. (2008) Biochemistry 47, 787–797 - PubMed
-
- Nishizuka Y. (1986) Science 233, 305–312 - PubMed
-
- Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Lührmann R., Barcia-Blanco M. A., Manley J. L. (1994) Nature 368, 119–124 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

