The teleost anatomy ontology: anatomical representation for the genomics age
- PMID: 20547776
- PMCID: PMC2885267
- DOI: 10.1093/sysbio/syq013
The teleost anatomy ontology: anatomical representation for the genomics age
Abstract
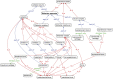
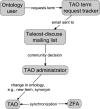
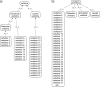
The rich knowledge of morphological variation among organisms reported in the systematic literature has remained in free-text format, impractical for use in large-scale synthetic phylogenetic work. This noncomputable format has also precluded linkage to the large knowledgebase of genomic, genetic, developmental, and phenotype data in model organism databases. We have undertaken an effort to prototype a curated, ontology-based evolutionary morphology database that maps to these genetic databases (http://kb.phenoscape.org) to facilitate investigation into the mechanistic basis and evolution of phenotypic diversity. Among the first requirements in establishing this database was the development of a multispecies anatomy ontology with the goal of capturing anatomical data in a systematic and computable manner. An ontology is a formal representation of a set of concepts with defined relationships between those concepts. Multispecies anatomy ontologies in particular are an efficient way to represent the diversity of morphological structures in a clade of organisms, but they present challenges in their development relative to single-species anatomy ontologies. Here, we describe the Teleost Anatomy Ontology (TAO), a multispecies anatomy ontology for teleost fishes derived from the Zebrafish Anatomical Ontology (ZFA) for the purpose of annotating varying morphological features across species. To facilitate interoperability with other anatomy ontologies, TAO uses the Common Anatomy Reference Ontology as a template for its upper level nodes, and TAO and ZFA are synchronized, with zebrafish terms specified as subtypes of teleost terms. We found that the details of ontology architecture have ramifications for querying, and we present general challenges in developing a multispecies anatomy ontology, including refinement of definitions, taxon-specific relationships among terms, and representation of taxonomically variable developmental pathways.
Figures
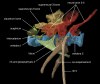
References
-
- Avraham S, Tung C-W, Ilic K, Jaiswal P, Kellogg E, McCouch S, Pujar A, Reiser L, Rhee S, Sachs M, Schaeffer M, Stein L, Stevens P, Leszek V, Zapata F, Ware D. The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res. 2008;36:D449–D454. - PMC - PubMed
-
- Bard JBL, Malone J, Rayner TF, Parkinson H. Proceedings of the 11th Annual Bio-Ontologies Meeting, 2008 July 20. Toronto: Canada; 2008. Minimal Anatomy Terminology (MAT): a species-independent terminology for anatomical mapping and retrieval.
-
- Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. Bgee: integrating and comparing heterogeneous transcriptome data among species. In: Bairoch A, Cohen-Boulakia S, Froidevaux C, editors. Data Integration in the Life Sciences, 5th International Workshop, DILS 2008, Evry France. Berlin: Springer-Verlag; 2008.
-
- Bird NC, Mabee PM. The developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae) Dev. Dyn. 2003;228:337–357. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

