Species delimitation using a combined coalescent and information-theoretic approach: an example from North American Myotis bats
- PMID: 20547777
- PMCID: PMC2885268
- DOI: 10.1093/sysbio/syq024
Species delimitation using a combined coalescent and information-theoretic approach: an example from North American Myotis bats
Abstract
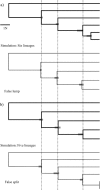
Coalescent model-based methods for phylogeny estimation force systematists to confront issues related to the identification of species boundaries. Unlike conventional phylogenetic analysis, where species membership can be assessed qualitatively after the phylogeny is estimated, the phylogenies that are estimated under a coalescent model treat aggregates of individuals as the operational taxonomic units and thus require a priori definition of these sets because the models assume that the alleles in a given lineage are sampled from a single panmictic population. Fortunately, the use of coalescent model-based approaches allows systematists to conduct probabilistic tests of species limits by calculating the probability of competing models of lineage composition. Here, we conduct the first exploration of the issues related to applying such tests to a complex empirical system. Sequence data from multiple loci were used to assess species limits and phylogeny in a clade of North American Myotis bats. After estimating gene trees at each locus, the likelihood of models representing all hierarchical permutations of lineage composition was calculated and Akaike information criterion scores were computed. Metrics borrowed from information theory suggest that there is strong support for several models that include multiple evolutionary lineages within the currently described species Myotis lucifugus and M. evotis. Although these results are preliminary, they illustrate the practical importance of coupled species delimitation and phylogeny estimation.
Figures
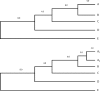
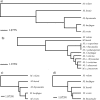
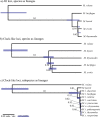
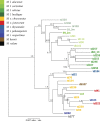
References
-
- Akaike H. Information theory as an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second International Symposium on Information Theory. Budapest (Hungary): Akademiai Kiado; 1973. pp. 267–281.
-
- Anderson DR. Model based inference in the life sciences. New York: Springer; 2008.
-
- Ané C, Larget B, Baum DA, Smith SD, Rokas A. Bayesian estimation of concordance among gene trees. Mol. Biol. Evol. 2007;24:412–426. - PubMed
-
- Baker RH, Stains HJ. A new long-eared Myotis (Myotis evotis) from northeastern Mexico. Univ. Kansas Mus. Nat. Hist. 1955;9:81–84.

