Structure-activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H
- PMID: 20547794
- PMCID: PMC2935023
- DOI: 10.1128/AAC.00434-10
Structure-activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H
Abstract
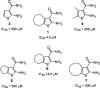
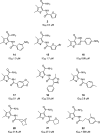
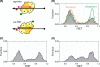
Vinylogous ureas 2-amino-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophene-3-carboxamide and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (compounds 1 and 2, respectively) were recently identified to be modestly potent inhibitors of the RNase H activity of HIV-1 and HIV-2 reverse transcriptase (RT). Both compounds shared a 3-CONH(2)-substituted thiophene ring but were otherwise structurally unrelated, which prevented a precise definition of the pharmacophore. We have therefore examined a larger series of vinylogous ureas carrying amide, amine, and cycloalkane modifications of the thiophene ring of compound 1. While cycloheptane- and cyclohexane-substituted derivatives retained potency, cyclopentane and cyclooctane substitutions eliminated activity. In the presence of a cycloheptane ring, modifying the 2-NH(2) or 3-CONH(2) functions decreased the potency. With respect to compound 2, vinylogous ureas whose dimethylthiophene ring contained modifications of the 2-NH(2) and 3-CONH(2) functions were investigated. 2-NH(2)-modified analogs displayed potency equivalent to or enhanced over that of compound 2, the most active of which, compound 16, reflected intramolecular cyclization of the 2-NH(2) and 3-CONH(2) groups. Molecular modeling was used to define an inhibitor binding site in the p51 thumb subdomain, suggesting that an interaction with the catalytically conserved His539 of the p66 RNase H domain could underlie inhibition of RNase H activity. Collectively, our data indicate that multiple functional groups of vinylogous ureas contribute to their potencies as RNase H inhibitors. Finally, single-molecule spectroscopy indicates that vinylogous ureas have the property of altering the reverse transcriptase orientation on a model RNA-DNA hybrid mimicking initiation plus-strand DNA synthesis.
Figures





Similar articles
-
Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity.ACS Chem Biol. 2008 Oct 17;3(10):635-44. doi: 10.1021/cb8001039. Epub 2008 Oct 3. ACS Chem Biol. 2008. PMID: 18831589 Free PMC article.
-
Mutagenesis of human immunodeficiency virus reverse transcriptase p51 subunit defines residues contributing to vinylogous urea inhibition of ribonuclease H activity.J Biol Chem. 2012 Feb 3;287(6):4066-75. doi: 10.1074/jbc.M111.314781. Epub 2011 Nov 21. J Biol Chem. 2012. PMID: 22105069 Free PMC article.
-
Synthesis, activity, and structural analysis of novel α-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H.J Med Chem. 2011 Jul 14;54(13):4462-73. doi: 10.1021/jm2000757. Epub 2011 Jun 2. J Med Chem. 2011. PMID: 21568335 Free PMC article.
-
Cutting into the Substrate Dominance: Pharmacophore and Structure-Based Approaches toward Inhibiting Human Immunodeficiency Virus Reverse Transcriptase-Associated Ribonuclease H.Acc Chem Res. 2020 Jan 21;53(1):218-230. doi: 10.1021/acs.accounts.9b00450. Epub 2019 Dec 27. Acc Chem Res. 2020. PMID: 31880912 Free PMC article. Review.
-
Recent progress in the research of small molecule HIV-1 RNase H inhibitors.Curr Med Chem. 2014 Jun;21(17):1956-67. doi: 10.2174/0929867321666140120121158. Curr Med Chem. 2014. PMID: 24438523 Review.
Cited by
-
The hepatitis B virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes.PLoS Pathog. 2013 Jan;9(1):e1003125. doi: 10.1371/journal.ppat.1003125. Epub 2013 Jan 22. PLoS Pathog. 2013. PMID: 23349632 Free PMC article.
-
Determinants of Active-Site Inhibitor Interaction with HIV-1 RNase H.ACS Infect Dis. 2019 Nov 8;5(11):1963-1974. doi: 10.1021/acsinfecdis.9b00300. Epub 2019 Oct 2. ACS Infect Dis. 2019. PMID: 31577424 Free PMC article.
-
Basic quinolinonyl diketo acid derivatives as inhibitors of HIV integrase and their activity against RNase H function of reverse transcriptase.J Med Chem. 2014 Apr 24;57(8):3223-34. doi: 10.1021/jm5001503. Epub 2014 Apr 11. J Med Chem. 2014. PMID: 24684270 Free PMC article.
-
The biology and synthesis of α-hydroxytropolones.Medchemcomm. 2014 Jul 1;5(7):842-852. doi: 10.1039/C4MD00055B. Medchemcomm. 2014. PMID: 25089179 Free PMC article.
-
dsRNA binding characterization of full length recombinant wild type and mutants Zaire ebolavirus VP35.Antiviral Res. 2012 Mar;93(3):354-63. doi: 10.1016/j.antiviral.2012.01.005. Epub 2012 Jan 25. Antiviral Res. 2012. PMID: 22289166 Free PMC article.
References
-
- Beard, W. A., S. J. Stahl, H. R. Kim, K. Bebenek, A. Kumar, M. P. Strub, S. P. Becerra, T. A. Kunkel, and S. H. Wilson. 1994. Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase. Alanine scanning mutagenesis of an alpha-helix in the thumb subdomain. J. Biol. Chem. 269:28091-28097. - PubMed
-
- Bonache, M. C., E. Quesada, C. W. Sheen, J. Balzarini, N. Sluis-Cremer, M. J. Perez-Perez, M. J. Camarasa, and A. San-Felix. 2008. Novel N-3 substituted TSAO-T derivatives: synthesis and anti-HIV-evaluation. Nucleosides Nucleotides Nucleic Acids 27:351-367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Medical

