Adaptive resistance to the "last hope" antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS
- PMID: 20547815
- PMCID: PMC2916309
- DOI: 10.1128/AAC.00242-10
Adaptive resistance to the "last hope" antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS
Abstract
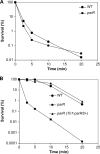
As multidrug resistance increases alarmingly, polymyxin B and colistin are increasingly being used in the clinic to treat serious Pseudomonas aeruginosa infections. In this opportunistic pathogen, subinhibitory levels of polymyxins and certain antimicrobial peptides induce resistance toward higher, otherwise lethal, levels of these antimicrobial agents. It is known that the modification of lipid A of lipopolysaccharide (LPS) is a key component of this adaptive peptide resistance, but to date, the regulatory mechanism underlying peptide regulation in P. aeruginosa has remained elusive. The PhoP-PhoQ and PmrA-PmrB two-component systems, which control this modification under low-Mg2+ conditions, do not appear to play a major role in peptide-mediated adaptive resistance, unlike in Salmonella where PhoQ is a peptide sensor. Here we describe the identification and characterization of a novel P. aeruginosa two-component regulator affecting polymyxin-adaptive resistance, ParR-ParS (PA1799-PA1798). This system was required for activation of the arnBCADTEF LPS modification operon in the presence of subinhibitory concentrations of polymyxin, colistin, or the bovine peptide indolicidin, leading to increased resistance to various polycationic antibiotics, including aminoglycosides. This study highlights the complexity of the regulatory network controlling resistance to cationic antibiotics and host peptides in P. aeruginosa, which has major relevance in the development and deployment of cationic antimicrobials.
Figures


References
-
- Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. - PubMed
-
- Bonomo, R. A., and D. Szabo. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43:SS49-SS56. - PubMed
-
- Choi, K. H., J. B. Gaynor, K. G. White, C. Lopez, C. M. Bosio, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443-448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

