Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells
- PMID: 20547861
- PMCID: PMC2900648
- DOI: 10.1073/pnas.0914143107
Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells
Abstract
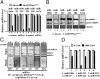
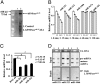
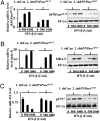
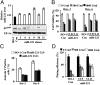
MicroRNAs (miRNA), small noncoding RNAs, affect a broad range of biological processes, including tumorigenesis, by targeting gene products that directly regulate cell growth. Human polynucleotide phosphorylase (hPNPase(old-35)), a type I IFN-inducible 3'-5' exoribonuclease, degrades specific mRNAs and small noncoding RNAs. The present study examined the effect of this enzyme on miRNA expression in human melanoma cells. miRNA microarray analysis of human melanoma cells infected with empty adenovirus or with an adenovirus expressing hPNPase(old-35) identified miRNAs differentially and specifically regulated by hPNPase(old-35). One of these, miR-221, a regulator of the cyclin-dependent kinase inhibitor p27(kip1), displayed robust down-regulation with ensuing up-regulation of p27(kip1) by expression of hPNPase(old-35), which also occurred in multiple human melanoma cells upon IFN-beta treatment. Using both in vivo immunoprecipitation followed by Northern blotting and RNA degradation assays, we confirm that mature miR-221 is the target of hPNPase(old-35). Inhibition of hPNPase(old-35) by shRNA or stable overexpression of miR-221 protected melanoma cells from IFN-beta-mediated growth inhibition, accentuating the importance of hPNPase(old-35) induction and miR-221 down-regulation in mediating IFN-beta action. Moreover, we now uncover a mechanism of miRNA regulation involving selective enzymatic degradation. Targeted overexpression of hPNPase(old-35) might provide an effective therapeutic strategy for miR-221-overexpressing and IFN-resistant tumors, such as melanoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Defining the mechanism by which IFN-beta dowregulates c-myc expression in human melanoma cells: pivotal role for human polynucleotide phosphorylase (hPNPaseold-35).Cell Death Differ. 2006 Sep;13(9):1541-53. doi: 10.1038/sj.cdd.4401829. Epub 2006 Jan 13. Cell Death Differ. 2006. PMID: 16410805
-
Progression elevated gene-3 promoter (PEG-Prom) confers cancer cell selectivity to human polynucleotide phosphorylase (hPNPase(old-35))-mediated growth suppression.J Cell Physiol. 2008 May;215(2):401-9. doi: 10.1002/jcp.21320. J Cell Physiol. 2008. PMID: 17960560
-
Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPase(old-35), a Type I interferon inducible early response gene.Gene. 2003 Oct 16;316:143-56. doi: 10.1016/s0378-1119(03)00752-2. Gene. 2003. PMID: 14563561
-
Human polynucleotide phosphorylase (hPNPase(old-35)): an evolutionary conserved gene with an expanding repertoire of RNA degradation functions.Oncogene. 2011 Apr 14;30(15):1733-43. doi: 10.1038/onc.2010.572. Epub 2010 Dec 13. Oncogene. 2011. PMID: 21151174 Free PMC article. Review.
-
Polynucleotide phosphorylase: an evolutionary conserved gene with an expanding repertoire of functions.Pharmacol Ther. 2006 Oct;112(1):243-63. doi: 10.1016/j.pharmthera.2006.04.003. Epub 2006 Jun 2. Pharmacol Ther. 2006. PMID: 16733069 Review.
Cited by
-
Mechanistic Actions of microRNAs in Diabetic Wound Healing.Cells. 2020 Oct 2;9(10):2228. doi: 10.3390/cells9102228. Cells. 2020. PMID: 33023156 Free PMC article. Review.
-
PNPase knockout results in mtDNA loss and an altered metabolic gene expression program.PLoS One. 2018 Jul 19;13(7):e0200925. doi: 10.1371/journal.pone.0200925. eCollection 2018. PLoS One. 2018. PMID: 30024931 Free PMC article.
-
Non-Coding RNA in Pancreas and β-Cell Development.Noncoding RNA. 2018 Dec 13;4(4):41. doi: 10.3390/ncrna4040041. Noncoding RNA. 2018. PMID: 30551650 Free PMC article. Review.
-
Role of miR-221/222 in Tumor Development and the Underlying Mechanism.J Oncol. 2019 Dec 24;2019:7252013. doi: 10.1155/2019/7252013. eCollection 2019. J Oncol. 2019. PMID: 31929798 Free PMC article.
-
Regulation of microRNA biogenesis.Nat Rev Mol Cell Biol. 2014 Aug;15(8):509-24. doi: 10.1038/nrm3838. Epub 2014 Jul 16. Nat Rev Mol Cell Biol. 2014. PMID: 25027649 Review.
References
-
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. - PubMed
-
- Tsuchiya S, Okuno Y, Tsujimoto G. MicroRNA: Biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J Pharmacol Sci. 2006;101:267–270. - PubMed
-
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. - PubMed
-
- Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

