Single-molecule spectroscopy of protein folding in a chaperonin cage
- PMID: 20547872
- PMCID: PMC2900638
- DOI: 10.1073/pnas.1002356107
Single-molecule spectroscopy of protein folding in a chaperonin cage
Abstract
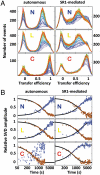
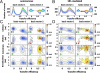
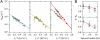
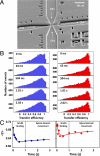
Molecular chaperones are known to be essential for avoiding protein aggregation in vivo, but it is still unclear how they affect protein folding mechanisms. We use single-molecule Förster resonance energy transfer to follow the folding of a protein inside the GroEL/GroES chaperonin cavity over a time range from milliseconds to hours. Our results show that confinement in the chaperonin decelerates the folding of the C-terminal domain in the substrate protein rhodanese, but leaves the folding rate of the N-terminal domain unaffected. Microfluidic mixing experiments indicate that strong interactions of the substrate with the cavity walls impede the folding process, but the folding hierarchy is preserved. Our results imply that no universal chaperonin mechanism exists. Rather, a competition between intra- and intermolecular interactions determines the folding rates and mechanisms of a substrate inside the GroEL/GroES cage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. - PubMed
-
- Hartl FU, Hayer-Hartl M. Protein folding—Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. - PubMed
-
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. - PubMed
-
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. - PubMed
-
- Fersht AR. Structure and mechanism in protein science. New York: W.H. Freeman and Company; 1998.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

