Regulation of innate immune responses by autophagy-related proteins
- PMID: 20548099
- PMCID: PMC2886348
- DOI: 10.1083/jcb.201002021
Regulation of innate immune responses by autophagy-related proteins
Abstract
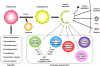
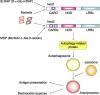
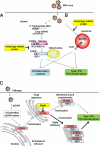
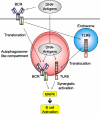
Pattern recognition receptors detect microbial components and induce innate immune responses, the first line of host defense against infectious agents. However, aberrant activation of immune responses often causes massive inflammation, leading to the development of autoimmune diseases. Therefore, both activation and inactivation of innate immune responses must be strictly controlled. Recent studies have shown that the cellular machinery associated with protein degradation, such as autophagy, is important for the regulation of innate immunity. These studies reveal that autophagy-related proteins are involved in the innate immune response and may contribute to the development of inflammatory disorders.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

