Molecular evolutionary analysis of the influenza A(H1N1)pdm, May-September, 2009: temporal and spatial spreading profile of the viruses in Japan
- PMID: 20548780
- PMCID: PMC2883557
- DOI: 10.1371/journal.pone.0011057
Molecular evolutionary analysis of the influenza A(H1N1)pdm, May-September, 2009: temporal and spatial spreading profile of the viruses in Japan
Abstract
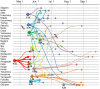
Background: In March 2009, pandemic influenza A(H1N1) (A(H1N1)pdm) emerged in Mexico and the United States. In Japan, since the first outbreak of A(H1N1)pdm in Osaka and Hyogo Prefectures occurred in the middle of May 2009, the virus had spread over 16 of 47 prefectures as of June 4, 2009.
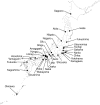
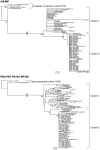
Methods/principal findings: We analyzed all-segment concatenated genome sequences of 75 isolates of A(H1N1)pdm viruses in Japan, and compared them with 163 full-genome sequences in the world. Two analyzing methods, distance-based and Bayesian coalescent MCMC inferences were adopted to elucidate an evolutionary relationship of the viruses in the world and Japan. Regardless of the method, the viruses in the world were classified into four distinct clusters with a few exceptions. Cluster 1 was originated earlier than cluster 2, while cluster 2 was more widely spread around the world. The other two clusters (clusters 1.2 and 1.3) were suggested to be distinct reassortants with different types of segment assortments. The viruses in Japan seemed to be a multiple origin, which were derived from approximately 28 transported cases. Twelve cases were associated with monophyletic groups consisting of Japanese viruses, which were referred to as micro-clade. While most of the micro-clades belonged to the cluster 2, the clade of the first cases of infection in Japan originated from cluster 1.2. Micro-clades of Osaka/Kobe and the Fukuoka cases, both of which were school-wide outbreaks, were eradicated. Time of most recent common ancestor (tMRCA) for each micro-clade demonstrated that some distinct viruses were transmitted in Japan between late May and early June, 2009, and appeared to spread nation-wide throughout summer.
Conclusions: Our results suggest that many viruses were transmitted from abroad in late May 2009 irrespective of preventive actions against the pandemic influenza, and that the influenza A(H1N1)pdm had become a pandemic stage in June 2009 in Japan.
Conflict of interest statement
Figures

Similar articles
-
Mutation analysis of 2009 pandemic influenza A(H1N1) viruses collected in Japan during the peak phase of the pandemic.PLoS One. 2011 Apr 29;6(4):e18956. doi: 10.1371/journal.pone.0018956. PLoS One. 2011. PMID: 21572517 Free PMC article.
-
Atypical influenza A(H1N1)pdm09 strains caused an influenza virus outbreak in Saudi Arabia during the 2009-2011 pandemic season.J Infect Public Health. 2019 Jul-Aug;12(4):557-567. doi: 10.1016/j.jiph.2019.01.067. Epub 2019 Feb 21. J Infect Public Health. 2019. PMID: 30799182
-
Whole genome characterization of human influenza A(H1N1)pdm09 viruses isolated from Kenya during the 2009 pandemic.Infect Genet Evol. 2016 Jun;40:98-103. doi: 10.1016/j.meegid.2016.02.029. Epub 2016 Feb 26. Infect Genet Evol. 2016. PMID: 26921801
-
Novel reassortant influenza viruses between pandemic (H1N1) 2009 and other influenza viruses pose a risk to public health.Microb Pathog. 2015 Dec;89:62-72. doi: 10.1016/j.micpath.2015.09.002. Epub 2015 Sep 4. Microb Pathog. 2015. PMID: 26344393 Review.
-
Evolutionary complexities of swine flu H1N1 gene sequences of 2009.Biochem Biophys Res Commun. 2009 Dec 18;390(3):349-51. doi: 10.1016/j.bbrc.2009.09.060. Epub 2009 Sep 19. Biochem Biophys Res Commun. 2009. PMID: 19769939 Review.
Cited by
-
Reconstruction of the evolutionary dynamics of the A(H1N1)pdm09 influenza virus in Italy during the pandemic and post-pandemic phases.PLoS One. 2012;7(11):e47517. doi: 10.1371/journal.pone.0047517. Epub 2012 Nov 9. PLoS One. 2012. PMID: 23152755 Free PMC article.
-
The high mutation rate at the D614G hotspot-furin cleavage site region increases the priming efficiency of the Spike protein by furin protease: analysis of Indonesian SARS-CoV-2 G614 variants obtained during the early COVID-19 pandemic.Virusdisease. 2023 May 31;34(2):1-10. doi: 10.1007/s13337-023-00827-w. Online ahead of print. Virusdisease. 2023. PMID: 37363361 Free PMC article.
-
Influenza risk management: lessons learned from an A(H1N1) pdm09 outbreak investigation in an operational military setting.PLoS One. 2013 Jul 10;8(7):e68639. doi: 10.1371/journal.pone.0068639. Print 2013. PLoS One. 2013. PMID: 23874699 Free PMC article.
-
Viral genetic sequence variations in pandemic H1N1/2009 and seasonal H3N2 influenza viruses within an individual, a household and a community.J Clin Virol. 2011 Oct;52(2):146-50. doi: 10.1016/j.jcv.2011.06.022. Epub 2011 Jul 29. J Clin Virol. 2011. PMID: 21802983 Free PMC article.
-
Virus evolution and transmission in an ever more connected world.Proc Biol Sci. 2015 Dec 22;282(1821):20142878. doi: 10.1098/rspb.2014.2878. Proc Biol Sci. 2015. PMID: 26702033 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention. Update: swine influenza A (H1N1) infections–California and Texas, April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:435–437. - PubMed
-
- Centers for Disease Control and Prevention. Outbreak of swine-origin influenza A (H1N1) virus infection - Mexico, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:467–470. - PubMed
-
- World Health Organization. Influenza A(H1N1)–Update 40. 2009. Available at http://www.who.int/csr/don/2009_06_12/en/index.html June 14:, in press.
-
- Ministry of Health LaW. Official notification [in Japanese]. 2009. Available: http://www.mhlw.go.jp/kunkyu/kenkou/influenza/090429-02.html via Internet. Accessed 29 April 2009.
-
- Shimada T, Gu Y, Kamiya H, Komiya N, Odaira F, et al. Epidemiology of influenza A(H1N1)v virus infection in Japan, May–June 2009. Eurosurveillance. 2009;14:19244. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

