Molecular analysis of malassezia microflora on the skin of the patients with atopic dermatitis
- PMID: 20548879
- PMCID: PMC2883395
- DOI: 10.5021/ad.2010.22.1.41
Molecular analysis of malassezia microflora on the skin of the patients with atopic dermatitis
Abstract
Background: The yeasts of the genus Malassezia are members of the normal flora on human skin and they are found in 75~80% of healthy adults. Since its association with various skin disorders have been known, there have been a growing number of reports that have implicated Malassezia yeast in atopic dermatitis (AD).
Objective: The aim of the present study is to isolate the various Malassezia species from AD patients by using 26S rDNA (ribosomal Deoxyribonucleic acid) PCR-RFLP and to investigate the relationship between a positive Malassezia culture and the severity of AD.
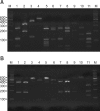
Methods: Cultures for Malassezia yeasts were taken from the scalp, cheek, chest, arm and thigh of 60 patients with atopic dermatitis. We used a rapid and accurate molecular biological method 26S rDNA PCR-RFLP, and this method can overcome the limits of the morphological and biochemical methods.
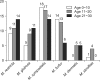
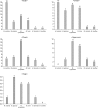
Results: Positive Malassezia growth was noted on 51.7% of the patients with atopic dermatitis by 26S rDNA PCR-RFLP analysis. The overall dominant species was M. sympodialis (16.3%). M. restricta was the most common species on the scalp (30.0%) and cheek (16.7%). M. sympodialis (28.3%) was the most common species on the chest. The positive culture rate was the highest for the 11~20 age group (59.0%) and the scalp showed the highest rate at 66.7%. There was no significant relationship between the Malassezia species and SCORing for Atopic Dermatitis (SCORAD).
Conclusion: The fact that the cultured species was different for the atopic dermatitis lesion skin from that of the normal skin may be due to the disrupted skin barrier function and sensitization of the organism induced by scratching in the AD lesion-skin. But there was no relationship between the Malassezia type and the severity score. The severity score is thought to depend not on the type, but also on the quantity of the yeast.
Keywords: 26S rDNA PCR-RFLP; Atopic dermatitis; Malassezia.
Figures
Similar articles
-
The Investigation on the Distribution of Malassezia Yeasts on the Normal Korean Skin by 26S rDNA PCR-RFLP.Ann Dermatol. 2009 Feb;21(1):18-26. doi: 10.5021/ad.2009.21.1.18. Epub 2009 Feb 28. Ann Dermatol. 2009. PMID: 20548850 Free PMC article.
-
Epidemiologic Study of Malassezia Yeasts in Seborrheic Dermatitis Patients by the Analysis of 26S rDNA PCR-RFLP.Ann Dermatol. 2010 May;22(2):149-55. doi: 10.5021/ad.2010.22.2.149. Epub 2010 May 17. Ann Dermatol. 2010. PMID: 20548904 Free PMC article.
-
Distribution of Malassezia species on the skin of patients with atopic dermatitis, psoriasis, and healthy volunteers assessed by conventional and molecular identification methods.BMC Dermatol. 2014 Mar 7;14:3. doi: 10.1186/1471-5945-14-3. BMC Dermatol. 2014. PMID: 24602368 Free PMC article.
-
[Malassezia species in patients with seborrheic dermatitis and atopic dermatitis].Nihon Ishinkin Gakkai Zasshi. 2005;46(3):163-7. doi: 10.3314/jjmm.46.163. Nihon Ishinkin Gakkai Zasshi. 2005. PMID: 16094289 Review. Japanese.
-
Malassezia species in healthy skin and in dermatological conditions.Int J Dermatol. 2016 May;55(5):494-504. doi: 10.1111/ijd.13116. Epub 2015 Dec 29. Int J Dermatol. 2016. PMID: 26710919 Review.
Cited by
-
The Malassezia genus in skin and systemic diseases.Clin Microbiol Rev. 2012 Jan;25(1):106-41. doi: 10.1128/CMR.00021-11. Clin Microbiol Rev. 2012. PMID: 22232373 Free PMC article. Review.
-
Head and neck dermatitis is exacerbated by Malassezia furfur colonization, skin barrier disruption, and immune dysregulation.Front Immunol. 2023 Feb 22;14:1114321. doi: 10.3389/fimmu.2023.1114321. eCollection 2023. Front Immunol. 2023. PMID: 36911720 Free PMC article.
-
Glucocorticosteroids and ciclosporin do not significantly impact canine cutaneous microbiota.BMC Vet Res. 2018 Feb 23;14(1):51. doi: 10.1186/s12917-018-1370-y. BMC Vet Res. 2018. PMID: 29471815 Free PMC article.
-
Epidemiology and changes in patient-related factors from 1997 to 2009 in clinical yeast isolates related to dermatology, gynaecology, and paediatrics.Int J Microbiol. 2013;2013:703905. doi: 10.1155/2013/703905. Epub 2013 Dec 11. Int J Microbiol. 2013. PMID: 24391669 Free PMC article.
-
A systematic literature review of the human skin microbiome as biomarker for dermatological drug development.Br J Clin Pharmacol. 2018 Oct;84(10):2178-2193. doi: 10.1111/bcp.13662. Epub 2018 Jul 19. Br J Clin Pharmacol. 2018. PMID: 29877593 Free PMC article.
References
-
- Ahn KJ. Taxonomy of the genus Malassezia. Korean J Med Mycol. 1998;3:81–88.
-
- Ljubojevic S, Skerlev M, Lipozencic J, Basta-Juzbasic A. The role of Malassezia furfur in dermatology. Clin Dermatol. 2002;20:179–182. - PubMed
-
- Nikkels AF, Pierard GE. Framing the future of antifungals in atopic dermatitis. Dermatology. 2003;206:398–400. - PubMed
LinkOut - more resources
Full Text Sources

