Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state
- PMID: 20548960
- PMCID: PMC2883604
- DOI: 10.1371/journal.pgen.1000982
Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state
Abstract
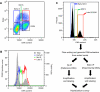
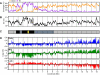
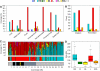
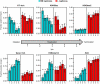
DNA replication programs have been studied extensively in yeast and animal systems, where they have been shown to correlate with gene expression and certain epigenetic modifications. Despite the conservation of core DNA replication proteins, little is known about replication programs in plants. We used flow cytometry and tiling microarrays to profile DNA replication of Arabidopsis thaliana chromosome 4 (chr4) during early, mid, and late S phase. Replication profiles for early and mid S phase were similar and encompassed the majority of the euchromatin. Late S phase exhibited a distinctly different profile that includes the remaining euchromatin and essentially all of the heterochromatin. Termination zones were consistent between experiments, allowing us to define 163 putative replicons on chr4 that clustered into larger domains of predominately early or late replication. Early-replicating sequences, especially the initiation zones of early replicons, displayed a pattern of epigenetic modifications specifying an open chromatin conformation. Late replicons, and the termination zones of early replicons, showed an opposite pattern. Histone H3 acetylated on lysine 56 (H3K56ac) was enriched in early replicons, as well as the initiation zones of both early and late replicons. H3K56ac was also associated with expressed genes, but this effect was local whereas replication time correlated with H3K56ac over broad regions. The similarity of the replication profiles for early and mid S phase cells indicates that replication origin activation in euchromatin is stochastic. Replicon organization in Arabidopsis is strongly influenced by epigenetic modifications to histones and DNA. The domain organization of Arabidopsis is more similar to that in Drosophila than that in mammals, which may reflect genome size and complexity. The distinct patterns of association of H3K56ac with gene expression and early replication provide evidence that H3K56ac may be associated with initiation zones and replication origins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Genome-Wide Analysis of the Arabidopsis Replication Timing Program.Plant Physiol. 2018 Mar;176(3):2166-2185. doi: 10.1104/pp.17.01537. Epub 2018 Jan 4. Plant Physiol. 2018. PMID: 29301956 Free PMC article.
-
Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley.Chromosoma. 2001 May;110(2):83-92. doi: 10.1007/s004120100132. Chromosoma. 2001. PMID: 11453558
-
Studies of mammalian chromosome replication. II. Evidence for the existence of defined chromosome replicating units.Chromosoma. 1981;83(5):721-41. doi: 10.1007/BF00328530. Chromosoma. 1981. PMID: 7028418
-
Links between genome replication and chromatin landscapes.Plant J. 2015 Jul;83(1):38-51. doi: 10.1111/tpj.12847. Epub 2015 Apr 29. Plant J. 2015. PMID: 25847096 Review.
-
Heterochromatin in interphase nuclei of Arabidopsis thaliana.Chromosome Res. 2003;11(3):227-40. doi: 10.1023/a:1022835825899. Chromosome Res. 2003. PMID: 12769290 Review.
Cited by
-
Regulating DNA replication in plants.Cold Spring Harb Perspect Biol. 2012 Dec 1;4(12):a010140. doi: 10.1101/cshperspect.a010140. Cold Spring Harb Perspect Biol. 2012. PMID: 23209151 Free PMC article. Review.
-
Cell cycle reentry from the late S phase: implications from stem cell formation in the moss Physcomitrella patens.J Plant Res. 2015 May;128(3):399-405. doi: 10.1007/s10265-015-0713-z. Epub 2015 Mar 24. J Plant Res. 2015. PMID: 25801272 Review.
-
Chromatin and DNA replication.Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a010207. doi: 10.1101/cshperspect.a010207. Cold Spring Harb Perspect Biol. 2013. PMID: 23751185 Free PMC article. Review.
-
A Protocol for Genome-Wide Analysis of DNA Replication Timing in Intact Root Tips.Methods Mol Biol. 2022;2382:29-72. doi: 10.1007/978-1-0716-1744-1_3. Methods Mol Biol. 2022. PMID: 34705232
-
Genome-scale analysis of replication timing: from bench to bioinformatics.Nat Protoc. 2011 Jun;6(6):870-95. doi: 10.1038/nprot.2011.328. Epub 2011 Jun 2. Nat Protoc. 2011. PMID: 21637205 Free PMC article.
References
-
- Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. - PubMed
-
- Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. - PubMed
-
- Gondor A, Ohlsson R. Replication timing and epigenetic reprogramming of gene expression: a two-way relationship? Nat Rev Genet. 2009;10:269–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

