Genomic patterns of nucleotide diversity in divergent populations of U.S. weedy rice
- PMID: 20550656
- PMCID: PMC2898691
- DOI: 10.1186/1471-2148-10-180
Genomic patterns of nucleotide diversity in divergent populations of U.S. weedy rice
Abstract
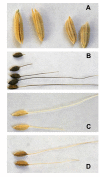
Background: Weedy rice (red rice), a conspecific weed of cultivated rice (Oryza sativa L.), is a significant problem throughout the world and an emerging threat in regions where it was previously absent. Despite belonging to the same species complex as domesticated rice and its wild relatives, the evolutionary origins of weedy rice remain unclear. We use genome-wide patterns of single nucleotide polymorphism (SNP) variation in a broad geographic sample of weedy, domesticated, and wild Oryza samples to infer the origin and demographic processes influencing U.S. weedy rice evolution.
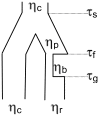
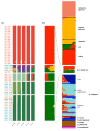
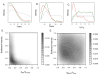
Results: We find greater population structure than has been previously reported for U.S. weedy rice, and that the multiple, genetically divergent populations have separate origins. The two main U.S. weedy rice populations share genetic backgrounds with cultivated O. sativa varietal groups not grown commercially in the U.S., suggesting weed origins from domesticated ancestors. Hybridization between weedy groups and between weedy rice and local crops has also led to the evolution of distinct U.S. weedy rice populations. Demographic simulations indicate differences among the main weedy groups in the impact of bottlenecks on their establishment in the U.S., and in the timing of divergence from their cultivated relatives.
Conclusions: Unlike prior research, we did not find unambiguous evidence for U.S. weedy rice originating via hybridization between cultivated and wild Oryza species. Our results demonstrate the potential for weedy life-histories to evolve directly from within domesticated lineages. The diverse origins of U.S. weedy rice populations demonstrate the multiplicity of evolutionary forces that can influence the emergence of weeds from a single species complex.
Figures

References
-
- Harlan JR. Crops and Man. Madison, Wisconsin: American Society of Agronomy; 1992.
-
- Ellstrand NC, Prentice HC, Hancock JF. Gene flow and introgression from domesticated plants into their wild relatives. Annual Review of Ecology and Systematics. 1999;10:539–563. doi: 10.1146/annurev.ecolsys.30.1.539. - DOI
-
- Holm LG, Plucknett DL, Pancho JV, Herberger JP. The world's worst weeds. Honolulu (USA): University Press of Hawaii; 1977.
-
- Dewet JMJ, Harlan JR. weeds and domesticates - evolution in man-made habitat. Econ Bot. 1975;10(2):99–107. doi: 10.1007/BF02863309. - DOI
-
- Gressel J. In: Crop Ferality and Volunteerism. J G, editor. CRC Press; 2005. Introduction: the challenges of ferality; pp. 1–9. full_text.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

