Does the core circadian clock in the moss Physcomitrella patens (Bryophyta) comprise a single loop?
- PMID: 20550695
- PMCID: PMC3017809
- DOI: 10.1186/1471-2229-10-109
Does the core circadian clock in the moss Physcomitrella patens (Bryophyta) comprise a single loop?
Abstract
Background: The endogenous circadian clock allows the organism to synchronize processes both to daily and seasonal changes. In plants, many metabolic processes such as photosynthesis, as well as photoperiodic responses, are under the control of a circadian clock. Comparative studies with the moss Physcomitrella patens provide the opportunity to study many aspects of land plant evolution. Here we present a comparative overview of clock-associated components and the circadian network in the moss P. patens.
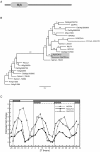
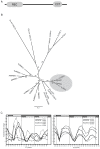
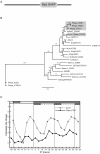
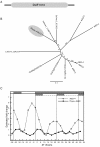
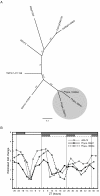
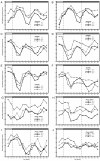
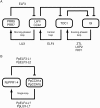
Results: The moss P. patens has a set of conserved circadian core components that share genetic relationship and gene expression patterns with clock genes of vascular plants. These genes include Myb-like transcription factors PpCCA1a and PpCCA1b, pseudo-response regulators PpPRR1-4, and regulatory elements PpELF3, PpLUX and possibly PpELF4. However, the moss lacks homologs of AtTOC1, AtGI and the AtZTL-family of genes, which can be found in all vascular plants studied here. These three genes constitute essential components of two of the three integrated feed-back loops in the current model of the Arabidopsis circadian clock mechanism. Consequently, our results suggest instead a single loop circadian clock in the moss. Possibly as a result of this, temperature compensation of core clock gene expression appears to be decreased in P. patens.
Conclusions: This study is the first comparative overview of the circadian clock mechanism in a basal land plant, the moss P. patens. Our results indicate that the moss clock mechanism may represent an ancestral state in contrast to the more complex and partly duplicated structure of subsequent land plants. These findings may provide insights into the understanding of the evolution of circadian network topology.
Figures
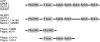
Similar articles
-
Functional characterization of CCA1/LHY homolog genes, PpCCA1a and PpCCA1b, in the moss Physcomitrella patens.Plant J. 2009 Nov;60(3):551-63. doi: 10.1111/j.1365-313X.2009.03979.x. Epub 2009 Jul 16. Plant J. 2009. PMID: 19624471
-
Photoperiod-dependent regulation of cell growth by PpCCA1a and PpCCA1b genes encoding single-myb clock proteins in the moss Physcomitrella patens.Genes Genet Syst. 2009 Oct;84(5):379-84. doi: 10.1266/ggs.84.379. Genes Genet Syst. 2009. PMID: 20154425
-
Heterologous expression and functional characterization of a Physcomitrella Pseudo response regulator homolog, PpPRR2, in Arabidopsis.Biosci Biotechnol Biochem. 2011;75(4):786-9. doi: 10.1271/bbb.100859. Epub 2011 Apr 22. Biosci Biotechnol Biochem. 2011. PMID: 21512222
-
Architecture of the PPR gene family in the moss Physcomitrella patens.RNA Biol. 2013;10(9):1439-45. doi: 10.4161/rna.24772. Epub 2013 Apr 23. RNA Biol. 2013. PMID: 23645116 Free PMC article. Review.
-
MicroRNAs in the moss Physcomitrella patens.Plant Mol Biol. 2012 Sep;80(1):55-65. doi: 10.1007/s11103-011-9761-5. Epub 2011 Mar 4. Plant Mol Biol. 2012. PMID: 21373961 Review.
Cited by
-
Circadian rhythms have significant effects on leaf-to-canopy scale gas exchange under field conditions.Gigascience. 2016 Oct 20;5(1):43. doi: 10.1186/s13742-016-0149-y. Gigascience. 2016. PMID: 27765071 Free PMC article.
-
Patterns of nucleotide diversity at photoperiod related genes in Norway spruce [Picea abies (L.) Karst].PLoS One. 2014 May 8;9(5):e95306. doi: 10.1371/journal.pone.0095306. eCollection 2014. PLoS One. 2014. PMID: 24810273 Free PMC article.
-
Core circadian clock and light signaling genes brought into genetic linkage across the green lineage.Plant Physiol. 2022 Sep 28;190(2):1037-1056. doi: 10.1093/plphys/kiac276. Plant Physiol. 2022. PMID: 35674369 Free PMC article.
-
Evolution of PAS domains and PAS-containing genes in eukaryotes.Chromosoma. 2014 Aug;123(4):385-405. doi: 10.1007/s00412-014-0457-x. Epub 2014 Apr 4. Chromosoma. 2014. PMID: 24699836
-
Nyctinastic thallus movement in the liverwort Marchantia polymorpha is regulated by a circadian clock.Sci Rep. 2020 May 26;10(1):8658. doi: 10.1038/s41598-020-65372-8. Sci Rep. 2020. PMID: 32457350 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

