Protein interaction network topology uncovers melanogenesis regulatory network components within functional genomics datasets
- PMID: 20550706
- PMCID: PMC2904735
- DOI: 10.1186/1752-0509-4-84
Protein interaction network topology uncovers melanogenesis regulatory network components within functional genomics datasets
Abstract
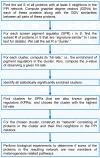
Background: RNA-mediated interference (RNAi)-based functional genomics is a systems-level approach to identify novel genes that control biological phenotypes. Existing computational approaches can identify individual genes from RNAi datasets that regulate a given biological process. However, currently available methods cannot identify which RNAi screen "hits" are novel components of well-characterized biological pathways known to regulate the interrogated phenotype. In this study, we describe a method to identify genes from RNAi datasets that are novel components of known biological pathways. We experimentally validate our approach in the context of a recently completed RNAi screen to identify novel regulators of melanogenesis.
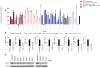
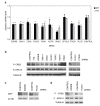
Results: In this study, we utilize a PPI network topology-based approach to identify targets within our RNAi dataset that may be components of known melanogenesis regulatory pathways. Our computational approach identifies a set of screen targets that cluster topologically in a human PPI network with the known pigment regulator Endothelin receptor type B (EDNRB). Validation studies reveal that these genes impact pigment production and EDNRB signaling in pigmented melanoma cells (MNT-1) and normal melanocytes.
Conclusions: We present an approach that identifies novel components of well-characterized biological pathways from functional genomics datasets that could not have been identified by existing statistical and computational approaches.
Figures
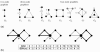

Similar articles
-
Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells.PLoS Genet. 2008 Dec;4(12):e1000298. doi: 10.1371/journal.pgen.1000298. Epub 2008 Dec 5. PLoS Genet. 2008. PMID: 19057677 Free PMC article.
-
A network-based integrative approach to prioritize reliable hits from multiple genome-wide RNAi screens in Drosophila.BMC Genomics. 2009 May 12;10:220. doi: 10.1186/1471-2164-10-220. BMC Genomics. 2009. PMID: 19435510 Free PMC article.
-
Integrated network analyses for functional genomic studies in cancer.Semin Cancer Biol. 2013 Aug;23(4):213-8. doi: 10.1016/j.semcancer.2013.06.004. Epub 2013 Jun 27. Semin Cancer Biol. 2013. PMID: 23811269 Free PMC article. Review.
-
Revealing molecular mechanisms by integrating high-dimensional functional screens with protein interaction data.PLoS Comput Biol. 2014 Sep 4;10(9):e1003801. doi: 10.1371/journal.pcbi.1003801. eCollection 2014 Sep. PLoS Comput Biol. 2014. PMID: 25188415 Free PMC article.
-
Functional genomics and proteomics of the cellular osmotic stress response in 'non-model' organisms.J Exp Biol. 2007 May;210(Pt 9):1593-601. doi: 10.1242/jeb.000141. J Exp Biol. 2007. PMID: 17449824 Review.
Cited by
-
GraphCrunch 2: Software tool for network modeling, alignment and clustering.BMC Bioinformatics. 2011 Jan 19;12:24. doi: 10.1186/1471-2105-12-24. BMC Bioinformatics. 2011. PMID: 21244715 Free PMC article.
-
Identification of human disease genes from interactome network using graphlet interaction.PLoS One. 2014 Jan 22;9(1):e86142. doi: 10.1371/journal.pone.0086142. eCollection 2014. PLoS One. 2014. PMID: 24465923 Free PMC article.
-
Estimating the activity of transcription factors by the effect on their target genes.Bioinformatics. 2014 Sep 1;30(17):i401-7. doi: 10.1093/bioinformatics/btu446. Bioinformatics. 2014. PMID: 25161226 Free PMC article.
-
Survey of network-based approaches to research of cardiovascular diseases.Biomed Res Int. 2014;2014:527029. doi: 10.1155/2014/527029. Epub 2014 Mar 20. Biomed Res Int. 2014. PMID: 24772427 Free PMC article. Review.
-
A molecular systems approach to modelling human skin pigmentation: identifying underlying pathways and critical components.BMC Res Notes. 2015 Apr 29;8:170. doi: 10.1186/s13104-015-1128-6. BMC Res Notes. 2015. PMID: 25925987 Free PMC article.
References
-
- Lamitina T. Functional genomic approaches in C. elegans. Methods Mol Biol. 2006;351:127–138. - PubMed

