Classification of G-protein coupled receptors based on support vector machine with maximum relevance minimum redundancy and genetic algorithm
- PMID: 20550715
- PMCID: PMC2905366
- DOI: 10.1186/1471-2105-11-325
Classification of G-protein coupled receptors based on support vector machine with maximum relevance minimum redundancy and genetic algorithm
Abstract
Background: Because a priori knowledge about function of G protein-coupled receptors (GPCRs) can provide useful information to pharmaceutical research, the determination of their function is a quite meaningful topic in protein science. However, with the rapid increase of GPCRs sequences entering into databanks, the gap between the number of known sequence and the number of known function is widening rapidly, and it is both time-consuming and expensive to determine their function based only on experimental techniques. Therefore, it is vitally significant to develop a computational method for quick and accurate classification of GPCRs.
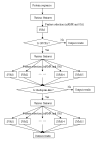
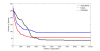
Results: In this study, a novel three-layer predictor based on support vector machine (SVM) and feature selection is developed for predicting and classifying GPCRs directly from amino acid sequence data. The maximum relevance minimum redundancy (mRMR) is applied to pre-evaluate features with discriminative information while genetic algorithm (GA) is utilized to find the optimized feature subsets. SVM is used for the construction of classification models. The overall accuracy with three-layer predictor at levels of superfamily, family and subfamily are obtained by cross-validation test on two non-redundant dataset. The results are about 0.5% to 16% higher than those of GPCR-CA and GPCRPred.
Conclusion: The results with high success rates indicate that the proposed predictor is a useful automated tool in predicting GPCRs. GPCR-SVMFS, a corresponding executable program for GPCRs prediction and classification, can be acquired freely on request from the authors.
Figures





Similar articles
-
GPCR-MPredictor: multi-level prediction of G protein-coupled receptors using genetic ensemble.Amino Acids. 2012 May;42(5):1809-23. doi: 10.1007/s00726-011-0902-6. Epub 2011 Apr 20. Amino Acids. 2012. PMID: 21505826
-
GPCRpred: an SVM-based method for prediction of families and subfamilies of G-protein coupled receptors.Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W383-9. doi: 10.1093/nar/gkh416. Nucleic Acids Res. 2004. PMID: 15215416 Free PMC article.
-
An improved classification of G-protein-coupled receptors using sequence-derived features.BMC Bioinformatics. 2010 Aug 9;11:420. doi: 10.1186/1471-2105-11-420. BMC Bioinformatics. 2010. PMID: 20696050 Free PMC article.
-
Present perspectives on the automated classification of the G-protein coupled receptors (GPCRs) at the protein sequence level.Curr Top Med Chem. 2011;11(15):1994-2009. doi: 10.2174/156802611796391221. Curr Top Med Chem. 2011. PMID: 21470173 Review.
-
Proteomic applications of automated GPCR classification.Proteomics. 2007 Aug;7(16):2800-14. doi: 10.1002/pmic.200700093. Proteomics. 2007. PMID: 17639603 Review.
Cited by
-
Emerging Role of Metabolomics in Ovarian Cancer Diagnosis.Metabolites. 2020 Oct 19;10(10):419. doi: 10.3390/metabo10100419. Metabolites. 2020. PMID: 33086611 Free PMC article. Review.
-
Accurate prediction of subcellular location of apoptosis proteins combining Chou's PseAAC and PsePSSM based on wavelet denoising.Oncotarget. 2017 Nov 21;8(64):107640-107665. doi: 10.18632/oncotarget.22585. eCollection 2017 Dec 8. Oncotarget. 2017. PMID: 29296195 Free PMC article.
-
Discriminating cirRNAs from other lncRNAs using a hierarchical extreme learning machine (H-ELM) algorithm with feature selection.Mol Genet Genomics. 2018 Feb;293(1):137-149. doi: 10.1007/s00438-017-1372-7. Epub 2017 Sep 14. Mol Genet Genomics. 2018. PMID: 28913654
-
The Use of Gene Ontology Term and KEGG Pathway Enrichment for Analysis of Drug Half-Life.PLoS One. 2016 Oct 25;11(10):e0165496. doi: 10.1371/journal.pone.0165496. eCollection 2016. PLoS One. 2016. PMID: 27780226 Free PMC article.
-
Computational Approach to Investigating Key GO Terms and KEGG Pathways Associated with CNV.Biomed Res Int. 2018 Apr 11;2018:8406857. doi: 10.1155/2018/8406857. eCollection 2018. Biomed Res Int. 2018. PMID: 29850576 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

