The mechanical properties of PCNA: implications for the loading and function of a DNA sliding clamp
- PMID: 20550919
- PMCID: PMC2884248
- DOI: 10.1016/j.bpj.2010.03.056
The mechanical properties of PCNA: implications for the loading and function of a DNA sliding clamp
Abstract
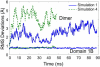
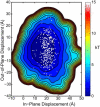
Sliding clamps are toroidal proteins that encircle DNA and act as mobile platforms for DNA replication and repair machinery. To be loaded onto DNA, the eukaryotic sliding clamp Proliferating Cell Nuclear Antigen (PCNA) must be splayed open at one of the subunit-subunit interfaces by the ATP-dependent clamp loader, Replication Factor C, whose clamp-interacting sites form a right-handed spiral. Earlier molecular dynamics (MD) studies suggested that when PCNA opens, it preferentially adopts a right-handed spiral to match the spiral of the clamp loader. Here, analysis of considerably longer MD simulations shows that although the opened form of PCNA can achieve conformations matching the helical pitch of Replication Factor C, it is not biased toward a right-handed spiral structure. A coarse-grained elastic model was also built; its strong correspondence to the all-atom MD simulations of PCNA suggests that the behavior of the open clamp is primarily due to elastic deformation governed by the topology of the clamp domains. The elastic model was further used to construct the energy landscape of the opened PCNA clamp, including conformations that would allow binding to the clamp loader and loading onto double-stranded DNA. A picture of PCNA emerges of a rather flexible protein that, once opened, is mechanically compliant in the clamp opening process.
(c) 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Kinetic analysis of PCNA clamp binding and release in the clamp loading reaction catalyzed by Saccharomyces cerevisiae replication factor C.Biochim Biophys Acta. 2015 Jan;1854(1):31-8. doi: 10.1016/j.bbapap.2014.09.019. Epub 2014 Oct 23. Biochim Biophys Acta. 2015. PMID: 25450506 Free PMC article.
-
A central swivel point in the RFC clamp loader controls PCNA opening and loading on DNA.J Mol Biol. 2012 Feb 17;416(2):163-75. doi: 10.1016/j.jmb.2011.12.017. Epub 2011 Dec 13. J Mol Biol. 2012. PMID: 22197374 Free PMC article.
-
Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex.Nature. 2004 Jun 17;429(6993):724-30. doi: 10.1038/nature02585. Nature. 2004. PMID: 15201901
-
The RFC clamp loader: structure and function.Subcell Biochem. 2012;62:259-79. doi: 10.1007/978-94-007-4572-8_14. Subcell Biochem. 2012. PMID: 22918590 Free PMC article. Review.
-
Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication.Adv Exp Med Biol. 2017;1042:135-162. doi: 10.1007/978-981-10-6955-0_7. Adv Exp Med Biol. 2017. PMID: 29357057 Review.
Cited by
-
Investigation of sliding DNA clamp dynamics by single-molecule fluorescence, mass spectrometry and structure-based modeling.Nucleic Acids Res. 2018 Apr 6;46(6):3103-3118. doi: 10.1093/nar/gky125. Nucleic Acids Res. 2018. PMID: 29529283 Free PMC article.
-
Linchpin DNA-binding residues serve as go/no-go controls in the replication factor C-catalyzed clamp-loading mechanism.J Biol Chem. 2017 Sep 22;292(38):15892-15906. doi: 10.1074/jbc.M117.798702. Epub 2017 Aug 14. J Biol Chem. 2017. PMID: 28808059 Free PMC article.
-
Expression of a novel peptide derived from PCNA damages DNA and reverses cisplatin resistance.Cancer Chemother Pharmacol. 2014 Nov;74(5):981-93. doi: 10.1007/s00280-014-2574-x. Epub 2014 Sep 5. Cancer Chemother Pharmacol. 2014. PMID: 25190177 Free PMC article.
-
Impact of individual proliferating cell nuclear antigen-DNA contacts on clamp loading and function on DNA.J Biol Chem. 2012 Oct 12;287(42):35370-35381. doi: 10.1074/jbc.M112.399071. Epub 2012 Aug 17. J Biol Chem. 2012. PMID: 22902629 Free PMC article.
-
Conformational analysis of processivity clamps in solution demonstrates that tertiary structure does not correlate with protein dynamics.Structure. 2014 Apr 8;22(4):572-581. doi: 10.1016/j.str.2014.02.001. Epub 2014 Mar 6. Structure. 2014. PMID: 24613485 Free PMC article.
References
-
- Garg P., Burgers P.M.J. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. - PubMed
-
- Hubscher U., Maga G., Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. - PubMed
-
- Maga G., Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 2003;116:3051–3060. - PubMed
-
- Moldovan G.-L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. - PubMed
-
- Krishna T.S., Kong X.P., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

