FACADE: a fast and sensitive algorithm for the segmentation and calling of high resolution array CGH data
- PMID: 20551132
- PMCID: PMC2926625
- DOI: 10.1093/nar/gkq548
FACADE: a fast and sensitive algorithm for the segmentation and calling of high resolution array CGH data
Abstract
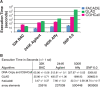
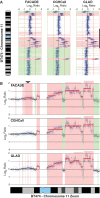
The availability of high resolution array comparative genomic hybridization (CGH) platforms has led to increasing complexities in data analysis. Specifically, defining contiguous regions of alterations or segmentation can be computationally intensive and popular algorithms can take hours to days for the processing of arrays comprised of hundreds of thousands to millions of elements. Additionally, tumors tend to demonstrate subtle copy number alterations due to heterogeneity, ploidy and hybridization effects. Thus, there is a need for fast, sensitive array CGH segmentation and alteration calling algorithms. Here, we describe Fast Algorithm for Calling After Detection of Edges (FACADE), a highly sensitive and easy to use algorithm designed to rapidly segment and call high resolution array data.
Figures

References
-
- Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007;89:647–653. - PubMed
-
- Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum. Mol. Genet. 2003;12(Spec No. 2):R145–R152. - PubMed
-
- Sagoo GS, Butterworth AS, Sanderson S, Shaw-Smith C, Higgins JP, Burton H. Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet. Med. 2009;11:139–146. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

