Evolutionary conservation of meiotic DSB proteins: more than just Spo11
- PMID: 20551169
- PMCID: PMC2885656
- DOI: 10.1101/gad.1944710
Evolutionary conservation of meiotic DSB proteins: more than just Spo11
Abstract
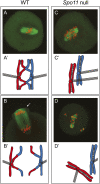
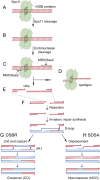
Meiotic recombination is initiated by programmed DNA double-strand breaks (DSBs) generated by the Spo11 protein. In budding yeast, five other meiotic-specific proteins are also required for DSB formation, but, with rare exception, orthologs had not been identified in other species. In this issue of Genes & Development, Kumar and colleagues (pp. 1266-1280) used a phylogenomic approach to identify two of these proteins across multiple clades, and confirmed that one of these, MEI4, is a functional ortholog in mouse.
Figures
Comment on
-
Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice.Genes Dev. 2010 Jun 15;24(12):1266-80. doi: 10.1101/gad.571710. Genes Dev. 2010. PMID: 20551173 Free PMC article.
References
-
- Arora C, Kee K, Maleki S, Keeney S 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell 13: 549–559 - PubMed
-
- Baudat F, de Massy B 2007. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577 - PubMed
-
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
