Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice
- PMID: 20551173
- PMCID: PMC2885662
- DOI: 10.1101/gad.571710
Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice
Abstract
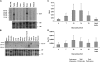
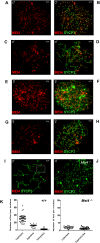
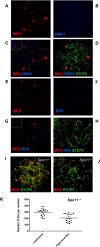
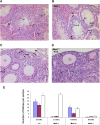
Meiotic recombination is initiated by the programmed induction of DNA double-strand breaks (DSBs) catalyzed by the evolutionarily conserved Spo11 protein. Studies in yeast have shown that DSB formation requires several other proteins, the role and conservation of which remain unknown. Here we show that two of these Saccharomyces cerevisiae proteins, Mei4 and Rec114, are evolutionarily conserved in most eukaryotes. Mei4(-/-) mice are deficient in meiotic DSB formation, thus showing the functional conservation of Mei4 in mice. Cytological analyses reveal that, in mice, MEI4 is localized in discrete foci on the axes of meiotic chromosomes that do not overlap with DMC1 and RPA foci. We thus propose that MEI4 acts as a structural component of the DSB machinery that ensures meiotic DSB formation on chromosome axes. We show that mouse MEI4 and REC114 proteins interact directly, and we identify conserved motifs as required for this interaction. Finally, the unexpected, concomitant absence of Mei4 and Rec114, as well as of Mnd1, Hop2, and Dmc1, in some eukaryotic species (particularly Neurospora crassa, Drosophila melanogaster, and Caenorhabditis elegans) suggests the existence of Mei4-Rec114-dependent and Mei4-Rec114-independent mechanisms for DSB formation, and a functional relationship between the chromosome axis and DSB formation.
Figures




Comment in
-
Evolutionary conservation of meiotic DSB proteins: more than just Spo11.Genes Dev. 2010 Jun 15;24(12):1201-7. doi: 10.1101/gad.1944710. Genes Dev. 2010. PMID: 20551169 Free PMC article.
References
-
- Arora C, Kee K, Maleki S, Keeney S 2004. Antiviral protein ski8 is a direct partner of spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell 13: 549–559 - PubMed
-
- Baudat F, de Massy B 2007. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577 - PubMed
-
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking spo11. Mol Cell 6: 989–998 - PubMed
-
- Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD 2005. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci 118: 3233–3245 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
