Negative allosteric modulators that target human alpha4beta2 neuronal nicotinic receptors
- PMID: 20551292
- PMCID: PMC2939664
- DOI: 10.1124/jpet.110.168211
Negative allosteric modulators that target human alpha4beta2 neuronal nicotinic receptors
Abstract
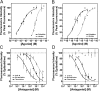
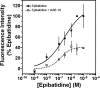
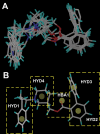
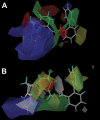
Allosteric modulation of neuronal nicotinic acetylcholine receptors (nAChRs) is considered to be one of the most promising approaches for therapeutics. We have previously reported on the pharmacological activity of several compounds that act as negative allosteric modulators (NAMs) of nAChRs. In the following studies, the effects of 30 NAMs from our small chemical library on both human alpha4beta2 (Halpha4beta2) and human alpha3beta4 (Halpha3beta4) nAChRs expressed in human embryonic kidney ts201 cells were investigated. During calcium accumulation assays, these NAMs inhibited nAChR activation with IC(50) values ranging from 2.4 microM to more than 100 microM. Several NAMs showed relative selectivity for Halpha4beta2 nAChRs with IC(50) values in the low micromolar range. A lead molecule, KAB-18, was identified that shows relative selectivity for Halpha4beta2 nAChRs. This molecule contains three phenyl rings, one piperidine ring, and one ester bond linkage. Structure-activity relationship (SAR) analyses of our data revealed three regions of KAB-18 that contribute to its relative selectivity. Predictive three-dimensional quantitative SAR (comparative molecular field analysis and comparative molecular similarity indices analysis) models were generated from these data, and a pharmacophore model was constructed to determine the chemical features that are important for biological activity. Using docking approaches and molecular dynamics on a Halpha4beta2 nAChR homology model, a binding mode for KAB-18 at the alpha/beta subunit interface that corresponds to the predicted pharmacophore is described. This binding mode was supported by mutagenesis studies. In summary, these studies highlight the importance of SAR, computational, and molecular biology approaches for the design and synthesis of potent and selective antagonists targeting specific nAChR subtypes.
Figures


References
-
- Anand R, Conroy WG, Schoepfer R, Whiting P, Lindstrom J. (1991) Neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes have a pentameric quaternary structure. J Biol Chem 266:11192–11198 - PubMed
-
- Arias HR. (1998) Binding sites for exogenous and endogenous non-competitive inhibitors of the nicotinic acetylcholine receptor. Biochim Biophys Acta 1376:173–220 - PubMed
-
- Bergmeier SC, Ismail KA, Arason KM, McKay S, Bryant DL, McKay DB. (2004) Structure activity studies of ring E analogues of methyllycaconitine. Part 2: synthesis of antagonists to the α3β4 nicotinic acetylcholine receptors through modifications to the ester. Bioorg Med Chem Lett 14:3739–3742 - PubMed
-
- Bergmeier SC, Lapinsky DJ, Free RB, McKay DB. (1999) Ring E analogs of methyllycaconitine (MLA) as novel nicotinic antagonists. Bioorg Med Chem Lett 9:2263–2266 - PubMed
-
- Bryant DL, Free RB, Thomasy SM, Lapinsky DJ, Ismail KA, McKay SB, Bergmeier SC, McKay DB. (2002) Structure-activity studies with ring E analogues of methyllycaconitine on bovine adrenal α3β4* nicotinic receptors. Neurosci Res 42:57–63 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

