Regulation of odor receptor genes in trichoid sensilla of the Drosophila antenna
- PMID: 20551440
- PMCID: PMC2940313
- DOI: 10.1534/genetics.110.117622
Regulation of odor receptor genes in trichoid sensilla of the Drosophila antenna
Abstract
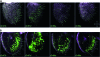
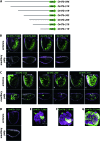
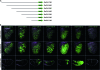
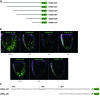
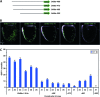
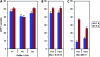
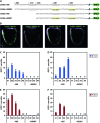
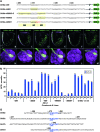
This study concerns the problem of odor receptor gene choice in the fruit fly Drosophila melanogaster. From a family of 60 Odor receptor genes, only one or a small number are selected for expression by each olfactory receptor neuron. Little is known about how an olfactory receptor neuron selects a receptor, or how the nucleotide sequences flanking a receptor gene dictate its expression in a particular neuron. Previous investigation has primarily concerned the maxillary palp, the simpler of the fly's two olfactory organs. Here we focus on genes encoding four antennal receptors that respond to fly odors in an in vivo expression system. To investigate the logic of odor receptor expression, we carry out a genetic analysis of their upstream regulatory sequences. Deletion analysis reveals that relatively short regulatory regions are sufficient to confer expression in the appropriate neurons, with limited if any misexpression. We find evidence for both positive and negative regulation. Multiple repressive functions restrict expression to the antenna, to a region of the antenna, and to neurons. Through deletion and base substitution mutagenesis we identify GCAATTA elements and find evidence that they act in both positive and negative regulation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

