Pollen-mediated gene flow in flax (Linum usitatissimum L.): can genetically engineered and organic flax coexist?
- PMID: 20551976
- PMCID: PMC3183899
- DOI: 10.1038/hdy.2010.81
Pollen-mediated gene flow in flax (Linum usitatissimum L.): can genetically engineered and organic flax coexist?
Abstract
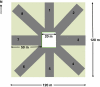
Coexistence allows growers and consumers the choice of producing or purchasing conventional or organic crops with known standards for adventitious presence of genetically engineered (GE) seed. Flax (Linum usitatissimum L.) is multipurpose oilseed crop in which product diversity and utility could be enhanced for industrial, nutraceutical and pharmaceutical markets through genetic engineering. If GE flax were released commercially, pollen-mediated gene flow will determine in part whether GE flax could coexist without compromising other markets. As a part of pre-commercialization risk assessment, we quantified pollen-mediated gene flow between two cultivars of flax. Field experiments were conducted at four locations during 2006 and 2007 in western Canada using a concentric donor (20 × 20 m) receptor (120 × 120 m) design. Gene flow was detected through the xenia effect of dominant alleles of high α-linolenic acid (ALA; 18:3(cisΔ9,12,15)) to the low ALA trait. Seeds were harvested from the pollen recipient plots up to a distance of 50 m in eight directions from the pollen donor. High ALA seeds were identified using a thiobarbituric acid test and served as a marker for gene flow. Binomial distribution and power analysis were used to predict the minimum number of seeds statistically required to detect the frequency of gene flow at specific α (confidence interval) and power (1-β) values. As a result of the low frequency of gene flow, approximately 4 million seeds were screened to derive accurate quantification. Frequency of gene flow was highest near the source: averaging 0.0185 at 0.1 m but declined rapidly with distance, 0.0013 and 0.00003 at 3 and 35 m, respectively. Gene flow was reduced to 50% (O₅₀) and 90% (O₉₀) between 0.85 to 2.64 m, and 5.68 to 17.56 m, respectively. No gene flow was detected at any site or year > 35 m distance from the pollen source, suggesting that frequency of gene flow was ≤ 0.00003 (P = 0.95). Although it is not possible to eliminate all adventitious presence caused by pollen-mediated gene flow, through harvest blending and the use of buffer zones between GE and conventional flax fields, it could be minimized. Managing other sources of adventitious presence including seed mixing and volunteer populations may be more problematic.
Figures

Comment in
-
Does flax have the answer to the GM mix-up?Heredity (Edinb). 2011 Jun;106(6):907-8. doi: 10.1038/hdy.2010.135. Epub 2010 Nov 10. Heredity (Edinb). 2011. PMID: 21063437 Free PMC article. No abstract available.
Similar articles
-
Heterologous expression of flax PHOSPHOLIPID:DIACYLGLYCEROL CHOLINEPHOSPHOTRANSFERASE (PDCT) increases polyunsaturated fatty acid content in yeast and Arabidopsis seeds.BMC Biotechnol. 2015 Jun 30;15:63. doi: 10.1186/s12896-015-0156-6. BMC Biotechnol. 2015. PMID: 26123542 Free PMC article.
-
Pollen-mediated gene flow from transgenic safflower (Carthamus tinctorius L.) intended for plant molecular farming to conventional safflower.Environ Biosafety Res. 2009 Jan-Mar;8(1):19-32. doi: 10.1051/ebr/2008023. Epub 2009 Feb 11. Environ Biosafety Res. 2009. PMID: 19419651
-
Pollen-mediated gene flow in wheat (Triticum aestivum L.) in a semiarid field environment in Spain.Transgenic Res. 2012 Dec;21(6):1329-39. doi: 10.1007/s11248-012-9619-x. Epub 2012 May 22. Transgenic Res. 2012. PMID: 22615061
-
Pollen-mediated intraspecific gene flow from herbicide resistant oilseed rape (Brassica napus L.).Transgenic Res. 2007 Oct;16(5):557-69. doi: 10.1007/s11248-007-9078-y. Epub 2007 May 31. Transgenic Res. 2007. PMID: 17541721 Review.
-
[Literature review of the dispersal of transgenes from genetically modified maize].C R Biol. 2009 Oct;332(10):861-75. doi: 10.1016/j.crvi.2009.07.001. Epub 2009 Aug 18. C R Biol. 2009. PMID: 19819407 Review. French.
Cited by
-
Rapid transgenerational adaptation in response to intercropping reduces competition.Elife. 2022 Sep 13;11:e77577. doi: 10.7554/eLife.77577. Elife. 2022. PMID: 36097813 Free PMC article.
-
The LuWD40-1 gene encoding WD repeat protein regulates growth and pollen viability in flax (Linum Usitatissimum L.).PLoS One. 2013 Jul 30;8(7):e69124. doi: 10.1371/journal.pone.0069124. Print 2013. PLoS One. 2013. PMID: 23935935 Free PMC article.
-
Modeling pollen-mediated gene flow from glyphosate-resistant to -susceptible giant ragweed (Ambrosia trifida L.) under field conditions.Sci Rep. 2017 Dec 6;7(1):17067. doi: 10.1038/s41598-017-16737-z. Sci Rep. 2017. PMID: 29213093 Free PMC article.
-
Pollen-mediated gene flow from glyphosate-resistant common waterhemp (Amaranthus rudis Sauer): consequences for the dispersal of resistance genes.Sci Rep. 2017 Mar 22;7:44913. doi: 10.1038/srep44913. Sci Rep. 2017. PMID: 28327669 Free PMC article.
-
Molecular markers as a complementary tool in risk assessments: quantifying interspecific gene flow from triticale to spring wheat and durum wheat.Transgenic Res. 2013 Aug;22(4):767-78. doi: 10.1007/s11248-012-9683-2. Epub 2013 Feb 7. Transgenic Res. 2013. PMID: 23389776
References
-
- Bausell RB, Li YF. Power Analysis for Experimental Research: A Practical Guide for the Biological, Medical and Social Sciences. Cambridge University Press: West Nyack, NY; 2002.
-
- Beckie HJ, Hall LM. Simple to complex: modelling crop pollen-mediated gene flow. Plant Sci. 2008;175:615–628.
-
- Beckie HJ, Warwick SI, Nair H, Seguin-Swartz G. Gene flow in commercial fields of herbicide-resistant canola (Brassica napus L.) Ecol Appl. 2003;13:1276–1294.
-
- Bedard D.2009Deregistered GM flax pops up in Europe [on-line]Available at http://www.albertafarmexpress.ca/issues/ISArticle.asp?aid=1000340642&PC ... (accessed 18 September 2009)Alberta Business Communication: Winnipeg, MB, Canada
-
- Bhatty RS, Rowland GG. Measurement of alpha-linolenic acid in the development of edible oil flax. J Am Oil Chem Soc. 1990;67:364–367.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

