Early reverse transcription is essential for productive foamy virus infection
- PMID: 20552014
- PMCID: PMC2884000
- DOI: 10.1371/journal.pone.0011023
Early reverse transcription is essential for productive foamy virus infection
Abstract
Background: Although viral RNA constitutes the majority of nucleic acids packaged in virions, a late occurring step of reverse transcription leads to the presence of infectious viral cDNA in foamy virus particles. This peculiarity distinguishes them from the rest of the retroviral family.
Principal findings: To evaluate the respective contribution of these viral nucleic acids in the replication of foamy viruses, their fate was studied by real-time PCR and RT-PCR early after infection, in the presence or in the absence of AZT. We found that an early reverse transcription step, which occurs during the first hours post-entry, is absolutely required for productive infection. Remarkably, sensitivity to AZT can be counteracted by increasing the multiplicity of infection (moi). We also show that 2-LTR circular viral DNA, which appears as soon as four hours post-infection, is transcriptionally competent.
Conclusion: Taken together, our data demonstrate that an early reverse transcription process, which takes place soon after viral entry, is indispensable for infectivity of FVs at low moi, when the amount of DNA-containing particles is not sufficient to lead to a productive infection. This study demonstrates a key role of the packaged viral RNA in the foamy virus infection, suggesting that the replication of this virus can be achieved by involving either viral DNA or RNA genome, depending on the condition of infection.
Conflict of interest statement
Figures
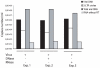
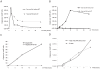



Similar articles
-
Evidence that the human foamy virus genome is DNA.J Virol. 1999 Feb;73(2):1565-72. doi: 10.1128/JVI.73.2.1565-1572.1999. J Virol. 1999. PMID: 9882362 Free PMC article.
-
Human foamy virus reverse transcription that occurs late in the viral replication cycle.J Virol. 1997 Oct;71(10):7305-11. doi: 10.1128/JVI.71.10.7305-7311.1997. J Virol. 1997. PMID: 9311807 Free PMC article.
-
Eleventh International Foamy Virus Conference-Meeting Report.Viruses. 2016 Nov 23;8(11):318. doi: 10.3390/v8110318. Viruses. 2016. PMID: 27886074 Free PMC article.
-
Why aren't foamy viruses pathogenic?Trends Microbiol. 2000 Jun;8(6):284-9. doi: 10.1016/s0966-842x(00)01763-7. Trends Microbiol. 2000. PMID: 10838587 Review.
-
Structural and Functional Aspects of Foamy Virus Protease-Reverse Transcriptase.Viruses. 2019 Jul 2;11(7):598. doi: 10.3390/v11070598. Viruses. 2019. PMID: 31269675 Free PMC article. Review.
Cited by
-
Foamy virus biology and its application for vector development.Viruses. 2011 May;3(5):561-85. doi: 10.3390/v3050561. Epub 2011 May 11. Viruses. 2011. PMID: 21994746 Free PMC article. Review.
-
Foamy virus assembly with emphasis on pol encapsidation.Viruses. 2013 Mar 20;5(3):886-900. doi: 10.3390/v5030886. Viruses. 2013. PMID: 23518575 Free PMC article. Review.
-
Expression of prototype foamy virus pol as a Gag-Pol fusion protein does not change the timing of reverse transcription.J Virol. 2013 Jan;87(2):1252-4. doi: 10.1128/JVI.02153-12. Epub 2012 Nov 7. J Virol. 2013. PMID: 23135709 Free PMC article.
-
SAMHD1 Functions and Human Diseases.Viruses. 2020 Mar 31;12(4):382. doi: 10.3390/v12040382. Viruses. 2020. PMID: 32244340 Free PMC article. Review.
-
Novel functions of prototype foamy virus Gag glycine- arginine-rich boxes in reverse transcription and particle morphogenesis.J Virol. 2011 Feb;85(4):1452-63. doi: 10.1128/JVI.01731-10. Epub 2010 Nov 24. J Virol. 2011. PMID: 21106749 Free PMC article.
References
-
- Delelis O, Carayon K, Guiot E, Leh H, Tauc P, et al. Insight into the integrase-DNA recognition mechanism. A specific DNA-binding mode revealed by an enzymatically labeled integrase. J Biol Chem. 2008;283:27838–27849. - PubMed
-
- Delelis O, Lehmann-Che J, Saib A. Foamy viruses–a world apart. Curr Opin Microbiol. 2004;7:400–406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

