A requirement for bid for induction of apoptosis by photodynamic therapy with a lysosome- but not a mitochondrion-targeted photosensitizer
- PMID: 20553412
- PMCID: PMC2974808
- DOI: 10.1111/j.1751-1097.2010.00766.x
A requirement for bid for induction of apoptosis by photodynamic therapy with a lysosome- but not a mitochondrion-targeted photosensitizer
Abstract
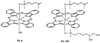
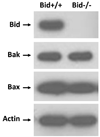
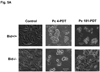
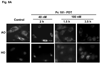
Photodynamic therapy (PDT) with lysosome-targeted photosensitizers induces the intrinsic pathway of apoptosis via the cleavage and activation of the BH3-only protein Bid by proteolytic enzymes released from photodisrupted lysosomes. To investigate the role of Bid in apoptosis induction and the role of damaged lysosomes on cell killing by lysosome-targeted PDT, we compared the responses of wild type and Bid-knock-out murine embryonic fibroblasts toward a mitochondrion/endoplasmic reticulum-binding photosensitizer, Pc 4, and a lysosome-targeted sensitizer, Pc 181. Whereas apoptosis and overall cell killing were induced equally well by Pc 4-PDT in both cell lines, Bid(-/-) cells were relatively resistant to induction of apoptosis and to overall killing following PDT with Pc 181, particularly at low PDT doses. Thus, Bid is critical for the induction of apoptosis caused by PDT with the lysosome-specific sensitizers, but dispensable for PDT targeted to other membranes.
© 2010 The Authors. Journal Compilation. The American Society of Photobiology.
Figures






















Similar articles
-
Hypericin-photodynamic therapy leads to interleukin-6 secretion by HepG2 cells and their apoptosis via recruitment of BH3 interacting-domain death agonist and caspases.Cell Death Dis. 2013 Jun 27;4(6):e697. doi: 10.1038/cddis.2013.219. Cell Death Dis. 2013. PMID: 23807226 Free PMC article.
-
Bid is required in NPe6-PDT-induced apoptosis.Photochem Photobiol. 2008 Jan-Feb;84(1):250-7. doi: 10.1111/j.1751-1097.2007.00248.x. Photochem Photobiol. 2008. PMID: 18173728
-
Lysosomal signaling enhances mitochondria-mediated photodynamic therapy in A431 cancer cells: role of iron.Photochem Photobiol. 2012 Mar-Apr;88(2):461-8. doi: 10.1111/j.1751-1097.2012.01081.x. Epub 2012 Jan 25. Photochem Photobiol. 2012. PMID: 22220628 Free PMC article.
-
Signaling pathways in cell death and survival after photodynamic therapy.J Photochem Photobiol B. 2000 Aug;57(1):1-13. doi: 10.1016/s1011-1344(00)00065-8. J Photochem Photobiol B. 2000. PMID: 11100832 Review.
-
Mitochondria-based photodynamic anti-cancer therapy.Adv Drug Deliv Rev. 2001 Jul 2;49(1-2):71-86. doi: 10.1016/s0169-409x(01)00126-0. Adv Drug Deliv Rev. 2001. PMID: 11377804 Review.
Cited by
-
Photodynamic Therapy and Immunity: An Update.Photochem Photobiol. 2020 May;96(3):550-559. doi: 10.1111/php.13253. Epub 2020 Apr 23. Photochem Photobiol. 2020. PMID: 32128821 Free PMC article. Review.
-
Effectiveness of ZnPc and of an amine derivative to inactivate Glioblastoma cells by Photodynamic Therapy: an in vitro comparative study.Sci Rep. 2019 Feb 28;9(1):3010. doi: 10.1038/s41598-019-39390-0. Sci Rep. 2019. PMID: 30816179 Free PMC article.
-
Apoptosis and associated phenomena as a determinants of the efficacy of photodynamic therapy.Photochem Photobiol Sci. 2015 Aug;14(8):1397-402. doi: 10.1039/c4pp00413b. Epub 2015 Jan 5. Photochem Photobiol Sci. 2015. PMID: 25559971 Free PMC article. Review.
-
Mitoferrin-2-dependent mitochondrial iron uptake sensitizes human head and neck squamous carcinoma cells to photodynamic therapy.J Biol Chem. 2013 Jan 4;288(1):677-86. doi: 10.1074/jbc.M112.422667. Epub 2012 Nov 7. J Biol Chem. 2013. PMID: 23135267 Free PMC article.
-
PDT-Induced Activation Enhanced by Hormone Response to Treatment.Int J Mol Sci. 2023 Sep 10;24(18):13917. doi: 10.3390/ijms241813917. Int J Mol Sci. 2023. PMID: 37762219 Free PMC article. Review.
References
-
- Moan J, Berg K. Photochemotherapy of cancer: experimental research. Photochem. Photobiol. 1992;55:931–948. - PubMed
-
- Weishaupt KR, Gomer CJ, Dougherty TJ. Identification of singlet oxygen as the cytotoxic agent in photoinactivation of a murine tumor. Cancer Res. 1976;36:2326–2329. - PubMed
-
- Peng Q, Moan J, Nesland JM. Correlation of subcellular and intratumoral photosensitizer localization with ultrastructural features after photodynamic therapy. Ultrastruct Pathol. 1996;20:109–129. - PubMed
-
- Oleinick NL, Morris RL, Belichenko I. The role of apoptosis in response to photodynamic therapy: what, where, why, and how. Photochem. Photobiol Sci. 2002;1:1–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

