Physical mapping of a large plant genome using global high-information-content-fingerprinting: the distal region of the wheat ancestor Aegilops tauschii chromosome 3DS
- PMID: 20553621
- PMCID: PMC2900270
- DOI: 10.1186/1471-2164-11-382
Physical mapping of a large plant genome using global high-information-content-fingerprinting: the distal region of the wheat ancestor Aegilops tauschii chromosome 3DS
Abstract
Background: Physical maps employing libraries of bacterial artificial chromosome (BAC) clones are essential for comparative genomics and sequencing of large and repetitive genomes such as those of the hexaploid bread wheat. The diploid ancestor of the D-genome of hexaploid wheat (Triticum aestivum), Aegilops tauschii, is used as a resource for wheat genomics. The barley diploid genome also provides a good model for the Triticeae and T. aestivum since it is only slightly larger than the ancestor wheat D genome. Gene co-linearity between the grasses can be exploited by extrapolating from rice and Brachypodium distachyon to Ae. tauschii or barley, and then to wheat.
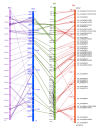
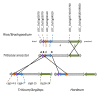
Results: We report the use of Ae. tauschii for the construction of the physical map of a large distal region of chromosome arm 3DS. A physical map of 25.4 Mb was constructed by anchoring BAC clones of Ae. tauschii with 85 EST on the Ae. tauschii and barley genetic maps. The 24 contigs were aligned to the rice and B. distachyon genomic sequences and a high density SNP genetic map of barley. As expected, the mapped region is highly collinear to the orthologous chromosome 1 in rice, chromosome 2 in B. distachyon and chromosome 3H in barley. However, the chromosome scale of the comparative maps presented provides new insights into grass genome organization. The disruptions of the Ae. tauschii-rice and Ae. tauschii-Brachypodium syntenies were identical. We observed chromosomal rearrangements between Ae. tauschii and barley. The comparison of Ae. tauschii physical and genetic maps showed that the recombination rate across the region dropped from 2.19 cM/Mb in the distal region to 0.09 cM/Mb in the proximal region. The size of the gaps between contigs was evaluated by comparing the recombination rate along the map with the local recombination rates calculated on single contigs.
Conclusions: The physical map reported here is the first physical map using fingerprinting of a complete Triticeae genome. This study demonstrates that global fingerprinting of the large plant genomes is a viable strategy for generating physical maps. Physical maps allow the description of the co-linearity between wheat and grass genomes and provide a powerful tool for positional cloning of new genes.
Figures

References
-
- Nesbitt M, Samuel D. In: 1st International Workshop on Hulled wheats. Padulosi S, Hammer K, Heller J, editor. Rome, Italy: International Plant Genetic Resources Institute; 1996. From staple crop to extinction? The archaeology and history of hulled wheats. Hulled Wheats. Promoting the conservation and use of underutilized and neglected crops; pp. 41–100.
-
- Kihara H. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric and Hort Tokyo. 1944;19:13–14.
-
- Dvorak J, Luo M, Yang Z, Zhang H. In: The Origins of Agriculture and Crop Domestication. Damania A, Valkoun J, Willcox G, Qualset C, editor. Aleppo, Syria: ICARDA; 1998. Genetic evidence on the origin of T. aestivum L; pp. 235–251.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

