Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs
- PMID: 20554048
- PMCID: PMC2974902
- DOI: 10.1016/j.ymeth.2010.06.008
Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs
Abstract
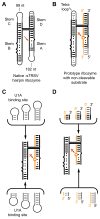
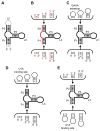
The structure determination of complex RNA molecules such as ribozymes, riboswitches and aptamers by X-ray crystallography hinges on the preparation of well-ordered crystals. Success usually results from molecular engineering to facilitate crystallization. An approach that has resulted in 10 new RNA structures in the past decade is the use of the U1A crystallization module. In this approach, the cognate site for the U1A spliceosomal protein is introduced into a functionally dispensable location in the RNA of interest, and the RNA is cocrystallized with the basic RNA-binding protein. In addition to facilitating crystallization, the presence of U1A can be useful for de novo phase determination. In this paper, some general considerations for the use of this approach to RNA crystallization are presented, and specifics of the application of the U1A module to the crystallization of the hairpin ribozyme and the tetracycline aptamer are reviewed.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Use of the U1A Protein to Facilitate Crystallization and Structure Determination of Large RNAs.Methods Mol Biol. 2016;1320:67-76. doi: 10.1007/978-1-4939-2763-0_6. Methods Mol Biol. 2016. PMID: 26227038 Free PMC article.
-
Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module.J Mol Biol. 2000 Jan 21;295(3):541-56. doi: 10.1006/jmbi.1999.3398. J Mol Biol. 2000. PMID: 10623545
-
Obtaining Crystals of Nucleic Acids in Complex with the Protein U1A Using the Soaking Method.Methods Mol Biol. 2022;2439:105-115. doi: 10.1007/978-1-0716-2047-2_8. Methods Mol Biol. 2022. PMID: 35226318
-
Two distinct catalytic strategies in the hepatitis δ virus ribozyme cleavage reaction.Biochemistry. 2011 Nov 8;50(44):9424-33. doi: 10.1021/bi201157t. Epub 2011 Oct 17. Biochemistry. 2011. PMID: 22003985 Free PMC article. Review.
-
RNA-protein interactions. Diverse modes of recognition.Curr Biol. 1995 Mar 1;5(3):249-51. doi: 10.1016/s0960-9822(95)00051-0. Curr Biol. 1995. PMID: 7540101 Review.
Cited by
-
Engineering RNA-binding proteins for biology.FEBS J. 2013 Aug;280(16):3734-54. doi: 10.1111/febs.12375. Epub 2013 Jul 5. FEBS J. 2013. PMID: 23742071 Free PMC article. Review.
-
Crystallographic analysis of TPP riboswitch binding by small-molecule ligands discovered through fragment-based drug discovery approaches.Methods Enzymol. 2014;549:221-33. doi: 10.1016/B978-0-12-801122-5.00010-6. Methods Enzymol. 2014. PMID: 25432751 Free PMC article.
-
RNA target highlights in CASP15: Evaluation of predicted models by structure providers.Proteins. 2023 Dec;91(12):1600-1615. doi: 10.1002/prot.26550. Epub 2023 Jul 19. Proteins. 2023. PMID: 37466021 Free PMC article.
-
An overview of biological macromolecule crystallization.Int J Mol Sci. 2013 May 31;14(6):11643-91. doi: 10.3390/ijms140611643. Int J Mol Sci. 2013. PMID: 23727935 Free PMC article. Review.
-
Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches.RNA. 2013 Feb;19(2):167-76. doi: 10.1261/rna.036269.112. Epub 2012 Dec 17. RNA. 2013. PMID: 23249744 Free PMC article.
References
-
- Drenth J. Principles of protein X-ray crystallography. Springer-Verlag; New York: 1994.
-
- Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AHJ, Seeman NC, Rich A. Science. 1974;185:435–40. - PubMed
-
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Nature. 1974;250:546–51. - PubMed
-
- Ferré-D’Amaré AR, Doudna JA. In: Current protocols in nucleic acid chemistry. Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. John Wiley & Sons; New York: 2000. pp. 7.6.1–7.6.10.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

