SHAPE-directed RNA secondary structure prediction
- PMID: 20554050
- PMCID: PMC2941709
- DOI: 10.1016/j.ymeth.2010.06.007
SHAPE-directed RNA secondary structure prediction
Abstract
The diverse functional roles of RNA are determined by its underlying structure. Accurate and comprehensive knowledge of RNA structure would inform a broader understanding of RNA biology and facilitate exploiting RNA as a biotechnological tool and therapeutic target. Determining the pattern of base pairing, or secondary structure, of RNA is a first step in these endeavors. Advances in experimental, computational, and comparative analysis approaches for analyzing secondary structure have yielded accurate structures for many small RNAs, but only a few large (>500 nts) RNAs. In addition, most current methods for determining a secondary structure require considerable effort, analytical expertise, and technical ingenuity. In this review, we outline an efficient strategy for developing accurate secondary structure models for RNAs of arbitrary length. This approach melds structural information obtained using SHAPE chemistry with structure prediction using nearest-neighbor rules and the dynamic programming algorithm implemented in the RNAstructure program. Prediction accuracies reach >or=95% for RNAs on the kilobase scale. This approach facilitates both development of new models and refinement of existing RNA structure models, which we illustrate using the Gag-Pol frameshift element in an HIV-1 M-group genome. Most promisingly, integrated experimental and computational refinement brings closer the ultimate goal of efficiently and accurately establishing the secondary structure for any RNA sequence.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
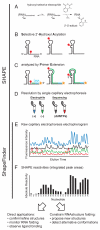

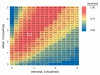
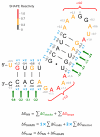
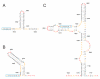
References
-
- Crothers DM, Cole PE, Hilbers CW, Shulman RG. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974;87:63–88. - PubMed
-
- Banerjee AR, Jaeger JA, Turner DH. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry. 1993;32:153–63. - PubMed
-
- Tinoco I, Jr., Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

