High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays
- PMID: 20554302
- PMCID: PMC2914195
- DOI: 10.1016/j.virol.2010.05.011
High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays
Abstract
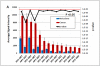
We have developed microarrays with all eight proteins encoded by 13 different human papillomavirus types associated with anogenital cancer (HPV-16, -18, -31, -33, -35, -45, and -53), genital warts (HPV-6 and -11), or skin lesions (HPV-1, -2, -4, and -5). We analyzed the seroprevalence of antibodies in 546 patients, which had either cervical carcinomas, or precursor lesions, or which were asymptomatic. All patient groups contained sera ranging from high reactivity against multiple HPV proteins to low or no reactivity. Computational analyses showed the E7 proteins of carcinogenic HPV types as significantly more reactive in cancer patients compared to asymptomatic individuals and discriminating between cancer and HSIL or LSIL patients. Antibodies against E4 and E5 had the highest seroprevalence but did not exhibit differential reactivity relative to pathology. Our study introduces a new approach to future evaluation of the overall antigenicity of HPV proteins and cross-reaction between homologous proteins.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures







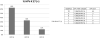
References
-
- Baker CC, Calef C. Maps of papillomavirus transcripts. In: Myers G, Bernard HU, Delius H, Baker CC, Icenogle J, Halpern A, Wheeler C, editors. Human papillomaviruses 1995 compendium. C. Los Alamos National Laboratory; Los Alamos, New Mexico: 1995. pp. 3–19. part III-A.
-
- Baldi P, Hatfield GW. Experiments to Data Analysis and Modeling. Cambridge University Press; 2002. DNA Microarrays and Gene Expression.
-
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. - PubMed
-
- Beare PA, Chen C, Bouman T, Pablo J, Unal B, Cockrell DC, Brown WC, Barbian KD, Porcella SF, Samuel JE, Felgner PL, Heinzen RA. Candidate antigens for Q fever serodiagnosis revealed by immunoscreening of a Coxiella burnetii protein microarray. Clin. Vaccine Immunol. 2008;15:1771–1779. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

