Closing the circle: replicating RNA with RNA
- PMID: 20554706
- PMCID: PMC2944364
- DOI: 10.1101/cshperspect.a002204
Closing the circle: replicating RNA with RNA
Abstract
How life emerged on this planet is one of the most important and fundamental questions of science. Although nearly all details concerning our origins have been lost in the depths of time, there is compelling evidence to suggest that the earliest life might have exploited the catalytic and self-recognition properties of RNA to survive. If an RNA based replicating system could be constructed in the laboratory, it would be much easier to understand the challenges associated with the very earliest steps in evolution and provide important insight into the establishment of the complex metabolic systems that now dominate this planet. Recent progress into the selection and characterization of ribozymes that promote nucleotide synthesis and RNA polymerization are discussed and outstanding problems in the field of RNA-mediated RNA replication are summarized.
Figures



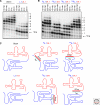
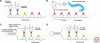
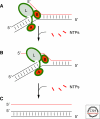
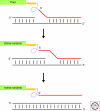
Similar articles
-
Co-operation between Polymerases and Nucleotide Synthetases in the RNA World.PLoS Comput Biol. 2016 Nov 7;12(11):e1005161. doi: 10.1371/journal.pcbi.1005161. eCollection 2016 Nov. PLoS Comput Biol. 2016. PMID: 27820829 Free PMC article.
-
Evolution in an RNA world.Cold Spring Harb Symp Quant Biol. 2009;74:17-23. doi: 10.1101/sqb.2009.74.004. Epub 2009 Aug 10. Cold Spring Harb Symp Quant Biol. 2009. PMID: 19667013 Free PMC article. Review.
-
The descent of polymerization.Nat Struct Biol. 2001 Jul;8(7):580-2. doi: 10.1038/89601. Nat Struct Biol. 2001. PMID: 11427883 No abstract available.
-
An RNA polymerase ribozyme that synthesizes its own ancestor.Proc Natl Acad Sci U S A. 2020 Feb 11;117(6):2906-2913. doi: 10.1073/pnas.1914282117. Epub 2020 Jan 27. Proc Natl Acad Sci U S A. 2020. PMID: 31988127 Free PMC article.
-
Re-creating an RNA world.Cell Mol Life Sci. 2006 Jun;63(11):1278-93. doi: 10.1007/s00018-006-6047-1. Cell Mol Life Sci. 2006. PMID: 16649141 Free PMC article. Review.
Cited by
-
Pragmatic turn in biology: From biological molecules to genetic content operators.World J Biol Chem. 2014 Aug 26;5(3):279-85. doi: 10.4331/wjbc.v5.i3.279. World J Biol Chem. 2014. PMID: 25225596 Free PMC article. Review.
-
Arginine cofactors on the polymerase ribozyme.PLoS One. 2011;6(9):e25030. doi: 10.1371/journal.pone.0025030. Epub 2011 Sep 20. PLoS One. 2011. PMID: 21949841 Free PMC article.
-
The paradox of dual roles in the RNA world: resolving the conflict between stable folding and templating ability.J Mol Evol. 2013 Sep;77(3):55-63. doi: 10.1007/s00239-013-9584-x. J Mol Evol. 2013. PMID: 24078151 Free PMC article.
-
Thermodynamic basis for the emergence of genomes during prebiotic evolution.PLoS Comput Biol. 2012 May;8(5):e1002534. doi: 10.1371/journal.pcbi.1002534. Epub 2012 May 31. PLoS Comput Biol. 2012. PMID: 22693440 Free PMC article.
-
Origin of Life: The Point of No Return.Life (Basel). 2020 Nov 3;10(11):269. doi: 10.3390/life10110269. Life (Basel). 2020. PMID: 33153087 Free PMC article.
References
-
- Bartel DP, Szostak JW 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418 - PubMed
-
- Bartel DP, Unrau PJ 1999. Constructing an RNA world. Trends Cell Biol 9: M9–M13 - PubMed
-
- Borukhov S, Nudler EC 2008. RNA polymerase: The vehicle of transcription. Trends Microbiol 16: 126–134 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
