Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses
- PMID: 20554771
- PMCID: PMC2919017
- DOI: 10.1128/JVI.02551-09
Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses
Abstract
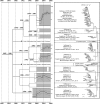
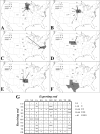
Type 2 (or North American-like) porcine reproductive and respiratory syndrome virus (PRRSV) was first recorded in 1987 in the United States and now occurs in most commercial swine industries throughout the world. In this study, we investigated the epidemiological and evolutionary behaviors of type 2 PRRSV. Based on phylogenetic analyses of 8,624 ORF5 sequences, we described a comprehensive picture of the diversity of type 2 PRRSVs and systematically classified all available sequences into lineages and sublineages, including a number of previously undescribed lineages. With the rapid growth of sequence deposition into the databases, it would be technically difficult for veterinary researchers to genotype their sequences by reanalyzing all sequences in the databases. To this end, a set of reference sequences was established based on our classification system, which represents the principal diversity of all available sequences and can readily be used for further genotyping studies. In addition, we further investigated the demographic histories of these lineages and sublineages by using Bayesian coalescence analyses, providing evolutionary insights into several important epidemiological events of type 2 PRRSV. Moreover, by using a phylogeographic approach, we were able to estimate the transmission frequencies between the pig-producing states in the United States and identified several states as the major sources of viral spread, i.e., "transmission centers." In summary, this study represents the most extensive phylogenetic analyses of type 2 PRRSV to date, providing a basis for future genotyping studies and dissecting the epidemiology of type 2 PRRSV from phylogenetic perspectives.
Figures



References
-
- An, T. Q., Y. J. Zhou, G. Q. Liu, Z. J. Tian, J. Li, H. J. Qiu, and G. Z. Tong. 2007. Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity. Vet. Microbiol. 123:43-52. - PubMed
-
- Andreyev, V. G., R. D. Wesley, W. L. Mengeling, A. C. Vorwald, and K. M. Lager. 1997. Genetic variation and phylogenetic relationships of 22 porcine reproductive and respiratory syndrome virus (PRRSV) field strains based on sequence analysis of open reading frame 5. Arch. Virol. 142:993-1001. - PubMed
-
- Bell, A. February. 1998. Hot PRRS is still hot. Pork 1998:32-33.
-
- Bush, E. J., B. Corso, J. Zimmerman, S. Swenson, D. Pyburn, and T. Burkgren. 1999. Update on the acute PRRS investigative study. Swine Health Prod. 7:179-180.
-
- Cai, H. Y., H. Alexander, S. Carman, D. Lloyd, G. Josephson, and M. G. Maxie. 2002. Restriction fragment length polymorphism of porcine reproductive and respiratory syndrome viruses recovered from Ontario farms, 1998-2000. J. Vet. Diagn. Invest. 14:343-347. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

