Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication
- PMID: 20554775
- PMCID: PMC2919037
- DOI: 10.1128/JVI.00508-10
Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication
Abstract
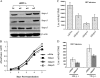
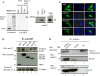
HIV-1 employs the cellular nuclear import machinery to actively transport its preintegration complex (PIC) into the nucleus for integration of the viral DNA. Several viral karyophilic proteins and cellular import factors have been suggested to contribute to HIV-1 PIC nuclear import and replication. However, how HIV interacts with different cellular machineries to ensure efficient nuclear import of its preintegration complex in dividing and nondividing cells is still not fully understood. In this study, we have investigated different importin alpha (Impalpha) family members for their impacts on HIV-1 replication, and we demonstrate that short hairpin RNA (shRNA)-mediated Impalpha3 knockdown (KD) significantly impaired HIV infection in HeLa cells, CD4(+) C8166 T cells, and primary macrophages. Moreover, quantitative real-time PCR analysis revealed that Impalpha3-KD resulted in significantly reduced levels of viral 2-long-terminal repeat (2-LTR) circles but had no effect on HIV reverse transcription. All of these data indicate an important role for Impalpha3 in HIV nuclear import. In an attempt to understand how Impalpha3 participates in HIV nuclear import and replication, we first demonstrated that the HIV-1 karyophilic protein integrase (IN) was able to interact with Impalpha3 both in a 293T cell expression system and in HIV-infected CD4(+) C8166 T cells. Deletion analysis suggested that a region (amino acids [aa] 250 to 270) in the C-terminal domain of IN is involved in this viral-cellular protein interaction. Overall, this study demonstrates for the first time that Impalpha3 is an HIV integrase-interacting cofactor that is required for efficient HIV-1 nuclear import and replication in both dividing and nondividing cells.
Figures
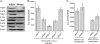

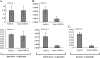

References
-
- Ao, Z., G. Huang, H. Yao, Z. Xu, M. Labine, A. W. Cochrane, and X. Yao. 2007. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 282:13456-13467. - PubMed
-
- Aratani, S., T. Oishi, H. Fujita, M. Nakazawa, R. Fujii, N. Imamoto, Y. Yoneda, A. Fukamizu, and T. Nakajima. 2006. The nuclear import of RNA helicase A is mediated by importin-α3. Biochem. Biophys. Res. Commun. 340:125-133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

