Fast nonnegative deconvolution for spike train inference from population calcium imaging
- PMID: 20554834
- PMCID: PMC3007657
- DOI: 10.1152/jn.01073.2009
Fast nonnegative deconvolution for spike train inference from population calcium imaging
Abstract
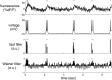
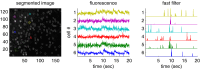
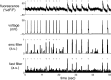
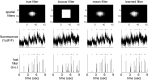
Fluorescent calcium indicators are becoming increasingly popular as a means for observing the spiking activity of large neuronal populations. Unfortunately, extracting the spike train of each neuron from a raw fluorescence movie is a nontrivial problem. This work presents a fast nonnegative deconvolution filter to infer the approximately most likely spike train of each neuron, given the fluorescence observations. This algorithm outperforms optimal linear deconvolution (Wiener filtering) on both simulated and biological data. The performance gains come from restricting the inferred spike trains to be positive (using an interior-point method), unlike the Wiener filter. The algorithm runs in linear time, and is fast enough that even when simultaneously imaging >100 neurons, inference can be performed on the set of all observed traces faster than real time. Performing optimal spatial filtering on the images further refines the inferred spike train estimates. Importantly, all the parameters required to perform the inference can be estimated using only the fluorescence data, obviating the need to perform joint electrophysiological and imaging calibration experiments.
Figures

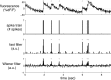
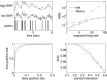
 (0, 2I) − 0.5
(0, 2I) − 0.5 (0, 2.5I), where
(0, 2.5I), where  (μ, Σ) indicates a 2-dimensional Gaussian with mean μ and covariance matrix Σ, β→ = 0, σ = 0.2, τ = 0.85 s, λ = 5 Hz, Δ = 5 ms, T = 1,200 time steps.
(μ, Σ) indicates a 2-dimensional Gaussian with mean μ and covariance matrix Σ, β→ = 0, σ = 0.2, τ = 0.85 s, λ = 5 Hz, Δ = 5 ms, T = 1,200 time steps.
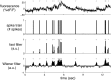
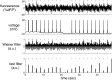
 ([−1, 0], 2I) − 0.5
([−1, 0], 2I) − 0.5 ([−1, 0], 2.5I), α→2 =
([−1, 0], 2.5I), α→2 =  ([1, 0], 2I) − 0.5
([1, 0], 2I) − 0.5 ([1, 0], 2.5I), β→ = 0, σ = 0.02, τ = 0.5 s, λ = 5 Hz, Δ = 5 ms, T = 1,200 time steps (not all time steps are shown).
([1, 0], 2.5I), β→ = 0, σ = 0.02, τ = 0.5 s, λ = 5 Hz, Δ = 5 ms, T = 1,200 time steps (not all time steps are shown).References
-
- Andrieu C, Barat É, Doucet A. Bayesian deconvolution of noisy filtered point processes. IEEE Trans Signal Process 49: 134–146, 2001
-
- Bell AJ, Sejnowski TJ. An information-maximisation approach to blind separation and blind deconvolution. Neural Comput 7: 1129–1159, 1995 - PubMed
-
- Boyd S, Vandenberghe L. Convex Optimization. Cambridge, UK: Cambridge Univ. Press, 2004
-
- Cover TM, Thomas JA. Elements of Information Theory: New York: Wiley–Interscience, 1991
-
- Cunningham JP, Shenoy KV, Sahani M. Fast Gaussian process methods for point process intensity estimation. In: Proceedings of the 25th International Conference on Machine Learning (ICML 2008) New York: IEEE Press, 2008, p. 192–199
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

