Airborne particulate matter selectively activates endoplasmic reticulum stress response in the lung and liver tissues
- PMID: 20554909
- PMCID: PMC2957267
- DOI: 10.1152/ajpcell.00529.2009
Airborne particulate matter selectively activates endoplasmic reticulum stress response in the lung and liver tissues
Abstract
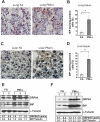
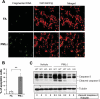
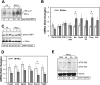
Recent studies have suggested a link between inhaled particulate matter (PM) exposure and increased mortality and morbidity associated with pulmonary and cardiovascular diseases. However, a precise understanding of the biological mechanism underlying PM-associated toxicity and pathogenesis remains elusive. Here, we investigated the impact of PM exposure in intracellular stress signaling pathways with animal models and cultured cells. Inhalation exposure of the mice to environmentally relevant fine particulate matter (aerodynamic diameter < 2.5 μm, PM(2.5)) induces endoplasmic reticulum (ER) stress and activation of unfolded protein response (UPR) in the lung and liver tissues as well as in the mouse macrophage cell line RAW264.7. Ambient PM(2.5) exposure activates double-strand RNA-activated protein kinase-like ER kinase (PERK), leading to phosphorylation of translation initiation factor eIF2α and induction of C/EBP homologous transcription factor CHOP/GADD153. Activation of PERK-mediated UPR pathway relies on the production of reactive oxygen species (ROS) and is critical for PM(2.5)-induced apoptosis. Furthermore, PM(2.5) exposure can activate ER stress sensor IRE1α, but it decreases the activity of IRE1α in splicing the mRNA encoding the UPR trans-activator X-box binding protein 1 (XBP1). Together, our study suggests that PM(2.5) exposure differentially activates the UPR branches, leading to ER stress-induced apoptosis through the PERK-eIF2α-CHOP UPR branch. This work provides novel insights into the cellular and molecular basis by which ambient PM(2.5) exposure elicits its cytotoxic effects that may be related to air pollution-associated pathogenesis.
Figures




Comment in
-
Endoplasmic reticulum stressed by pollution. Focus on "Airborne particulate matter selectively activates endoplasmic reticulum stress response in the lung and liver tissues".Am J Physiol Cell Physiol. 2010 Oct;299(4):C727-8. doi: 10.1152/ajpcell.00271.2010. Epub 2010 Jul 14. Am J Physiol Cell Physiol. 2010. PMID: 20631243 No abstract available.
Similar articles
-
Exposure to inhaled particulate matter activates early markers of oxidative stress, inflammation and unfolded protein response in rat striatum.Toxicol Lett. 2013 Oct 24;222(2):146-54. doi: 10.1016/j.toxlet.2013.07.012. Epub 2013 Jul 26. Toxicol Lett. 2013. PMID: 23892126 Free PMC article.
-
Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response.Cell Stress Chaperones. 2010 May;15(3):281-93. doi: 10.1007/s12192-009-0142-9. Epub 2009 Nov 8. Cell Stress Chaperones. 2010. PMID: 19898960 Free PMC article.
-
Selective, potent blockade of the IRE1 and ATF6 pathways by 4-phenylbutyric acid analogues.Br J Pharmacol. 2013 Oct;170(4):822-34. doi: 10.1111/bph.12306. Br J Pharmacol. 2013. PMID: 23869584 Free PMC article.
-
Endoplasmic reticulum stress in liver disease.J Hepatol. 2011 Apr;54(4):795-809. doi: 10.1016/j.jhep.2010.11.005. Epub 2010 Nov 13. J Hepatol. 2011. PMID: 21145844 Free PMC article. Review.
-
Endoplasmic reticulum stress and eIF2α phosphorylation: The Achilles heel of pancreatic β cells.Mol Metab. 2017 Jul 12;6(9):1024-1039. doi: 10.1016/j.molmet.2017.06.001. eCollection 2017 Sep. Mol Metab. 2017. PMID: 28951826 Free PMC article. Review.
Cited by
-
Endoplasmic reticulum stress as a pro-fibrotic stimulus.Biochim Biophys Acta. 2013 Jul;1832(7):940-7. doi: 10.1016/j.bbadis.2012.11.011. Epub 2012 Nov 28. Biochim Biophys Acta. 2013. PMID: 23201247 Free PMC article.
-
Asbestos-induced alveolar epithelial cell apoptosis. The role of endoplasmic reticulum stress response.Am J Respir Cell Mol Biol. 2013 Dec;49(6):892-901. doi: 10.1165/rcmb.2013-0053OC. Am J Respir Cell Mol Biol. 2013. PMID: 23885834 Free PMC article.
-
Organic compounds present in airborne particles stimulate superoxide production and DNA fragmentation: role of NOX and xanthine oxidase in animal tissues.Environ Sci Pollut Res Int. 2016 Aug;23(16):16653-60. doi: 10.1007/s11356-016-6833-8. Epub 2016 May 16. Environ Sci Pollut Res Int. 2016. PMID: 27180836
-
Non-Alcoholic Fatty Liver Disease: Metabolic, Genetic, Epigenetic and Environmental Risk Factors.Int J Environ Res Public Health. 2021 May 14;18(10):5227. doi: 10.3390/ijerph18105227. Int J Environ Res Public Health. 2021. PMID: 34069012 Free PMC article. Review.
-
A healthier approach to clinical trials evaluating resveratrol for primary prevention of age‐related diseases in healthy populations.Aging (Albany NY). 2013 Jul;5(7):495-506. doi: 10.18632/aging.100579. Aging (Albany NY). 2013. PMID: 24073437 Free PMC article.
References
-
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27: 53–66, 2007 - PubMed
-
- Arantes-Costa FM, Lopes FD, Toledo AC, Magliarelli-Filho PA, Moriya HT, Carvalho-Oliveira R, Mauad T, Saldiva PH, Martins MA. Effects of residual oil fly ash (ROFA) in mice with chronic allergic pulmonary inflammation. Toxicol Pathol 36: 680–686, 2008. - PubMed
-
- American Lung Association State of the Air 2009 http://www.lungusa2.org/sota/2009/SOTA-2009-Full-Print.pdf); 2008Highlights of Recent Research on Particulate Air Pollution: Effects of Long-Term Exposure (http://www.lungusa.org/site/c.dvLUK9O0E/b.36864/k.9F49/State_of_the_Air.htm).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

