Templating efficiency of naked DNA
- PMID: 20554916
- PMCID: PMC2901432
- DOI: 10.1073/pnas.0914872107
Templating efficiency of naked DNA
Abstract
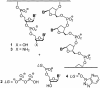
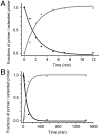
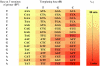
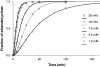
Template-directed synthesis of complementary strands is pivotal for life. Nature employs polymerases for this reaction, leaving the ability of DNA itself to direct the incorporation of individual nucleotides at the end of a growing primer difficult to assess. Using 64 sequences, we now find that any of the four nucleobases, in combination with any neighboring residue, support enzyme-free primer extension when primer and mononucleotide are sufficiently reactive, with >or=93% primer extension for all sequences. Between the 64 possible base triplets, the rate of extension for the poorest template, CAG, with A as templating base, and the most efficient template, TCT, with C as templating base, differs by less than two orders of magnitude. Further, primer extension with a balanced mixture of monomers shows >or=72% of the correct extension product in all cases, and >or=90% incorporation of the correct base for 46 out of 64 triplets in the presence of a downstream-binding strand. A mechanism is proposed with a binding equilibrium for the monomer, deprotonation of the primer, and two chemical steps, the first of which is most strongly modulated by the sequence. Overall, rates show a surprisingly smooth reactivity landscape, with similar incorporation on strongly and weakly templating sequences. These results help to clarify the substrate contribution to copying, as found in polymerase-catalyzed replication, and show an important feature of DNA as genetic material.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Chemical primer extension: individual steps of spontaneous replication.Chem Biodivers. 2007 Apr;4(4):784-802. doi: 10.1002/cbdv.200790064. Chem Biodivers. 2007. PMID: 17443889
-
The effect of leaving groups on binding and reactivity in enzyme-free copying of DNA and RNA.Nucleic Acids Res. 2016 Jul 8;44(12):5504-14. doi: 10.1093/nar/gkw476. Epub 2016 May 27. Nucleic Acids Res. 2016. PMID: 27235418 Free PMC article.
-
Tuning the reaction site for enzyme-free primer-extension reactions through small molecule substituents.Chemistry. 2006 Mar 8;12(9):2472-81. doi: 10.1002/chem.200501008. Chemistry. 2006. PMID: 16402399
-
Nucleotide insertion and primer extension at abasic template sites in different sequence contexts.Ann N Y Acad Sci. 1994 Jul 29;726:132-42; discussion 142-3. doi: 10.1111/j.1749-6632.1994.tb52804.x. Ann N Y Acad Sci. 1994. PMID: 8092671 Review.
-
Enzyme-free genetic copying of DNA and RNA sequences.Beilstein J Org Chem. 2018 Mar 12;14:603-617. doi: 10.3762/bjoc.14.47. eCollection 2018. Beilstein J Org Chem. 2018. PMID: 29623122 Free PMC article. Review.
Cited by
-
Evolutionary advantage of directional symmetry breaking in self-replicating polymers.J Theor Biol. 2018 Jun 7;446:128-136. doi: 10.1016/j.jtbi.2018.03.012. Epub 2018 Mar 12. J Theor Biol. 2018. PMID: 29544886 Free PMC article.
-
Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.Science. 2023 Oct 27;382(6669):423-429. doi: 10.1126/science.adh5339. Epub 2023 Oct 26. Science. 2023. PMID: 37883544 Free PMC article.
-
Synthesis of phosphoramidate-linked DNA by a modified DNA polymerase.Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7276-7283. doi: 10.1073/pnas.1922400117. Epub 2020 Mar 18. Proc Natl Acad Sci U S A. 2020. PMID: 32188786 Free PMC article.
-
What Does "the RNA World" Mean to "the Origin of Life"?Life (Basel). 2017 Nov 29;7(4):49. doi: 10.3390/life7040049. Life (Basel). 2017. PMID: 29186049 Free PMC article.
-
Synthesis of N3'-P5'-linked phosphoramidate DNA by nonenzymatic template-directed primer extension.J Am Chem Soc. 2013 Jan 16;135(2):924-32. doi: 10.1021/ja311164j. Epub 2013 Jan 7. J Am Chem Soc. 2013. PMID: 23252395 Free PMC article.
References
-
- Joyce C-M, Benkovic S-J. DNA polymerase fidelity: Kinetics structure and checkpoints. Biochemistry. 2004;43:14317–14324. - PubMed
-
- Berdis A-J. Mechanisms of DNA polymerases. Chem Rev. 2009;109:2862–2879. - PubMed
-
- Kunkel T-A, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

