RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma
- PMID: 20555022
- PMCID: PMC3130552
- DOI: 10.1126/scitranslmed.3000931
RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma
Abstract
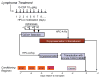
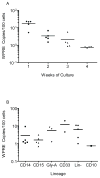
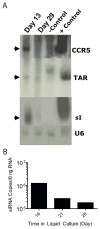
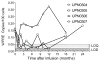
AIDS patients who develop lymphoma are often treated with transplanted hematopoietic progenitor cells. As a first step in developing a hematopoietic cell-based gene therapy treatment, four patients undergoing treatment with these transplanted cells were also given gene-modified peripheral blood-derived (CD34(+)) hematopoietic progenitor cells expressing three RNA-based anti-HIV moieties (tat/rev short hairpin RNA, TAR decoy, and CCR5 ribozyme). In vitro analysis of these gene-modified cells showed no differences in their hematopoietic potential compared with nontransduced cells. In vitro estimates of successful expression of the anti-HIV moieties were initially as high as 22% but declined to approximately 1% over 4 weeks of culture. Ethical study design required that patients be transplanted with both gene-modified and unmanipulated hematopoietic progenitor cells obtained from the patient by apheresis. Transfected cells were successfully engrafted in all four infused patients by day 11, and there were no unexpected infusion-related toxicities. Persistent vector expression in multiple cell lineages was observed at low levels for up to 24 months, as was expression of the introduced small interfering RNA and ribozyme. Therefore, we have demonstrated stable vector expression in human blood cells after transplantation of autologous gene-modified hematopoietic progenitor cells. These results support the development of an RNA-based cell therapy platform for HIV.
Figures
References
-
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science New York, NY. 2009;323:1304–1307. - PubMed
-
- Chen RY, Accortt NA, Westfall AO, Mugavero MJ, Raper JL, Cloud GA, Stone BK, Carter J, Call S, Pisu M, Allison J, Saag MS. Distribution of health care expenditures for HIV-infected patients. Clin Infect Dis. 2006;42:1003–1010. - PubMed
-
- Baltimore D. Gene therapy. Intracellular immunization. Nature. 1988;335:395–396. - PubMed
-
- Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI61839/AI/NIAID NIH HHS/United States
- R01 HL074704/HL/NHLBI NIH HHS/United States
- AI42552/AI/NIAID NIH HHS/United States
- R37 AI029329/AI/NIAID NIH HHS/United States
- HL07470/HL/NHLBI NIH HHS/United States
- M01 RR00043/RR/NCRR NIH HHS/United States
- P30 CA33572-26/CA/NCI NIH HHS/United States
- M01 RR000043/RR/NCRR NIH HHS/United States
- P01 AI061839/AI/NIAID NIH HHS/United States
- S10RR025083-01/RR/NCRR NIH HHS/United States
- P50 CA107399/CA/NCI NIH HHS/United States
- P30 CA033572/CA/NCI NIH HHS/United States
- R01 AI042552/AI/NIAID NIH HHS/United States
- S10 RR025083/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

