CLIA-tested genetic variants on commercial SNP arrays: potential for incidental findings in genome-wide association studies
- PMID: 20556870
- PMCID: PMC3056880
- DOI: 10.1097/GIM.0b013e3181e1e2a9
CLIA-tested genetic variants on commercial SNP arrays: potential for incidental findings in genome-wide association studies
Abstract
Purpose: Increases in throughput and affordability of genotyping products have led to large sample sizes in genetic studies, increasing the likelihood that incidental genetic findings may occur. We set out to survey potential notifiable variants on arrays used in genome-wide association studies and in direct-to-consumer genetic services.
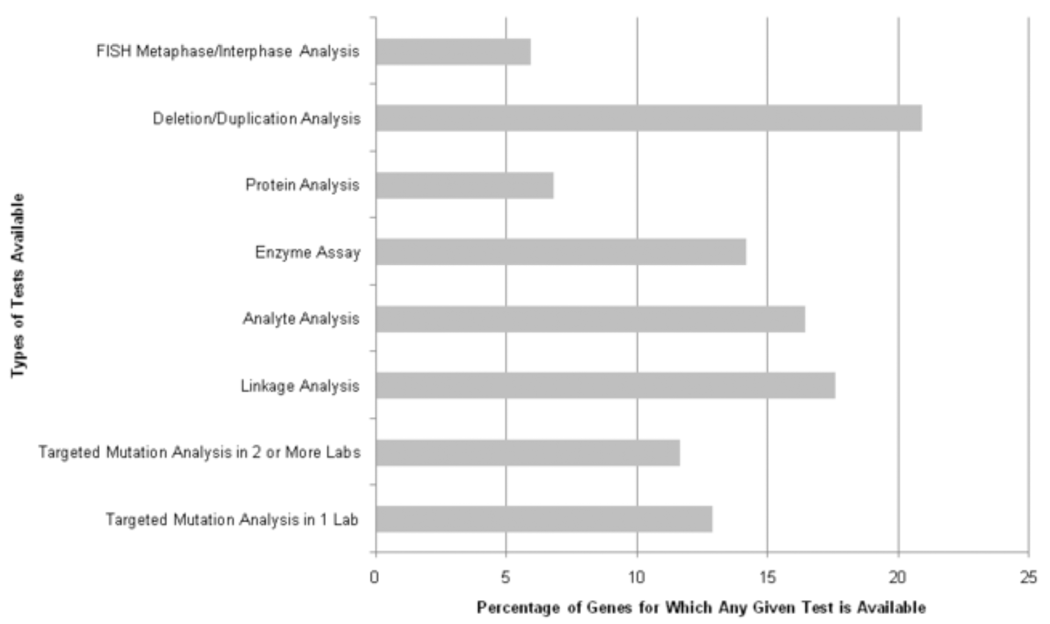
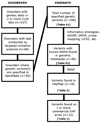
Methods: We used multiple bioinformatics strategies to identify, and map variants tested for genetic disorders in > or = 2 CLIA-approved laboratories (based on the GeneTests database). We subsequently surveyed 18 commercial single nucleotide polymorphism arrays and HapMap for these variants.
Results: Of 1,362 genes tested according to GeneTests, we identified 298 specific targeted mutations measured in more than or equal to two laboratories, encompassing 56 disorders. Only 88 of 298 mutations could be identified as known single nucleotide polymorphisms in genomic databases. We found 18 of 88 single nucleotide polymorphisms present in HapMap or on commercial single nucleotide polymorphism arrays. Homozygotes for rare alleles of some variants were identified in the Framingham Heart Study, an active genome-wide association studies cohort (n = 8,410).
Conclusions: Variants in genes including APOE, F5, HFE, CYP21A2, MEFV, SPINK1, BTD, GALT, and G6PD were found on single nucleotide polymorphism arrays or in the HapMap. Some of these variants may warrant further review to determine their likelihood to trigger incidental findings in the course of genome-wide association studies or direct-to-consumer testing.
Conflict of interest statement
None of the authors have a conflict of interest to disclose.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

