Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists
- PMID: 20558186
- PMCID: PMC2976827
- DOI: 10.1016/j.neuropharm.2010.06.002
Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists
Abstract
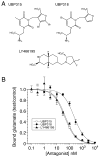
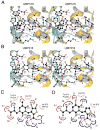
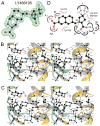
The availability of crystal structures for the ligand binding domains of ionotropic glutamate receptors, combined with their key role in synaptic function in the normal and diseased brain, offers a unique selection of targets for pharmaceutical research compared to other drug targets for which the atomic structure of the ligand binding site is not known. Currently only a few antagonist structures have been solved, and these reveal ligand specific conformational changes that hinder rational drug design. Here we report high resolution crystal structures for three kainate receptor GluK1 antagonist complexes which reveal new and unexpected modes of binding, highlighting the continued need for experimentally determined receptor-ligand complexes.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Lessons from crystal structures of kainate receptors.Neuropharmacology. 2017 Jan;112(Pt A):16-28. doi: 10.1016/j.neuropharm.2016.05.014. Epub 2016 May 26. Neuropharmacology. 2017. PMID: 27236079 Review.
-
Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.J Struct Biol. 2011 Dec;176(3):307-14. doi: 10.1016/j.jsb.2011.08.014. Epub 2011 Sep 1. J Struct Biol. 2011. PMID: 21907808
-
Structural and pharmacological characterization of phenylalanine-based AMPA receptor antagonists at kainate receptors.ChemMedChem. 2012 Oct;7(10):1793-8. doi: 10.1002/cmdc.201100599. Epub 2012 Mar 7. ChemMedChem. 2012. PMID: 22407805
-
Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists.J Neurosci. 2006 Mar 15;26(11):2852-61. doi: 10.1523/JNEUROSCI.0123-06.2005. J Neurosci. 2006. PMID: 16540562 Free PMC article.
-
Medicinal chemistry of competitive kainate receptor antagonists.ACS Chem Neurosci. 2011 Feb 16;2(2):60-74. doi: 10.1021/cn1001039. Epub 2010 Dec 10. ACS Chem Neurosci. 2011. PMID: 22778857 Free PMC article. Review.
Cited by
-
Structure, Function, and Regulation of the Kainate Receptor.Subcell Biochem. 2022;99:317-350. doi: 10.1007/978-3-031-00793-4_10. Subcell Biochem. 2022. PMID: 36151381 Review.
-
Novel Functional Properties of Drosophila CNS Glutamate Receptors.Neuron. 2016 Dec 7;92(5):1036-1048. doi: 10.1016/j.neuron.2016.10.058. Epub 2016 Nov 23. Neuron. 2016. PMID: 27889096 Free PMC article.
-
Kainate Receptor Antagonists: Recent Advances and Therapeutic Perspective.Int J Mol Sci. 2023 Jan 18;24(3):1908. doi: 10.3390/ijms24031908. Int J Mol Sci. 2023. PMID: 36768227 Free PMC article. Review.
-
Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain.Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):5921-6. doi: 10.1073/pnas.1217549110. Epub 2013 Mar 25. Proc Natl Acad Sci U S A. 2013. PMID: 23530186 Free PMC article.
-
Structural mechanism of glutamate receptor activation and desensitization.Nature. 2014 Oct 16;514(7522):328-34. doi: 10.1038/nature13603. Epub 2014 Aug 3. Nature. 2014. PMID: 25119039 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. - PMC - PubMed
-
- Alt A, Weiss B, Ornstein PL, Gleason SD, Bleakman D, Stratford RE, Jr, Witkin JM. Anxiolytic-like effects through a GLUK5 kainate receptor mechanism. Neuropharmacology. 2007;52:1482–1487. - PubMed
-
- Arinaminpathy Y, Sansom MS, Biggin PC. Binding site flexibility: Molecular simulation of partial and full agonists with a glutamate receptor. Mol Pharmacol. 2006;69:5–12. - PubMed
-
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

