Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA
- PMID: 20558719
- PMCID: PMC3075239
- DOI: 10.1126/science.1187197
Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA
Erratum in
- Science. 2010 Sep 17;329(5998):1467. Steitz, Joan [corrected to Steitz, Joan A]
Abstract
T cells transformed by Herpesvirus saimiri express seven viral U-rich noncoding RNAs of unknown function called HSURs. We noted that conserved sequences in HSURs 1 and 2 constitute potential binding sites for three host-cell microRNAs (miRNAs). Coimmunoprecipitation experiments confirmed that HSURs 1 and 2 interact with the predicted miRNAs in virally transformed T cells. The abundance of one of these miRNAs, miR-27, is dramatically lowered in transformed cells, with consequent effects on the expression of miR-27 target genes. Transient knockdown and ectopic expression of HSUR 1 demonstrate that it directs degradation of mature miR-27 in a sequence-specific and binding-dependent manner. This viral strategy illustrates use of a ncRNA to manipulate host-cell gene expression via the miRNA pathway.
Figures
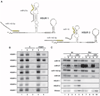

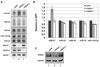

Comment in
-
Molecular biology. Paring miRNAs through pairing.Science. 2010 Jun 18;328(5985):1494-5. doi: 10.1126/science.1191531. Science. 2010. PMID: 20558697 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

