Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae
- PMID: 20558724
- PMCID: PMC2930708
- DOI: 10.1074/jbc.M109.071126
Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae
Abstract
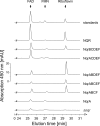
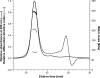
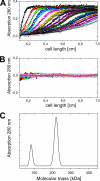
The sodium ion-translocating NADH:quinone oxidoreductase (Na(+)-NQR) from the human pathogen Vibrio cholerae is a respiratory membrane protein complex that couples the oxidation of NADH to the transport of Na(+) across the bacterial membrane. The Na(+)-NQR comprises the six subunits NqrABCDEF, but the stoichiometry and arrangement of these subunits are unknown. Redox-active cofactors are FAD and a 2Fe-2S cluster on NqrF, covalently attached FMNs on NqrB and NqrC, and riboflavin and ubiquinone-8 with unknown localization in the complex. By analyzing the cofactor content and NADH oxidation activity of subcomplexes of the Na(+)-NQR lacking individual subunits, the riboflavin cofactor was unequivocally assigned to the membrane-bound NqrB subunit. Quantitative analysis of the N-terminal amino acids of the holo-complex revealed that NqrB is present in a single copy in the holo-complex. It is concluded that the hydrophobic NqrB harbors one riboflavin in addition to its covalently attached FMN. The catalytic role of two flavins in subunit NqrB during the reduction of ubiquinone to ubiquinol by the Na(+)-NQR is discussed.
Figures






Similar articles
-
The single NqrB and NqrC subunits in the Na(+)-translocating NADH: quinone oxidoreductase (Na(+)-NQR) from Vibrio cholerae each carry one covalently attached FMN.Biochim Biophys Acta. 2012 Oct;1817(10):1817-22. doi: 10.1016/j.bbabio.2012.02.012. Epub 2012 Feb 16. Biochim Biophys Acta. 2012. PMID: 22366169
-
Riboflavin is an active redox cofactor in the Na+-pumping NADH: quinone oxidoreductase (Na+-NQR) from Vibrio cholerae.J Biol Chem. 2008 Nov 28;283(48):33162-7. doi: 10.1074/jbc.M806913200. Epub 2008 Oct 2. J Biol Chem. 2008. PMID: 18832377 Free PMC article.
-
The Electron Transfer Pathway of the Na+-pumping NADH:Quinone Oxidoreductase from Vibrio cholerae.J Biol Chem. 2009 Mar 27;284(13):8963-72. doi: 10.1074/jbc.M809395200. Epub 2009 Jan 20. J Biol Chem. 2009. PMID: 19155212 Free PMC article.
-
The Na+-translocating NADH:quinone oxidoreductase (Na+-NQR): Physiological role, structure and function of a redox-driven, molecular machine.Biochim Biophys Acta Bioenerg. 2024 Nov 1;1865(4):149485. doi: 10.1016/j.bbabio.2024.149485. Epub 2024 Jun 30. Biochim Biophys Acta Bioenerg. 2024. PMID: 38955304 Review.
-
The structure of Na⁺-translocating of NADH:ubiquinone oxidoreductase of Vibrio cholerae: implications on coupling between electron transfer and Na⁺ transport.Biol Chem. 2015 Sep;396(9-10):1015-30. doi: 10.1515/hsz-2015-0128. Biol Chem. 2015. PMID: 26146127 Review.
Cited by
-
Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes.mBio. 2013 Feb 12;4(1):e00615-12. doi: 10.1128/mBio.00615-12. mBio. 2013. PMID: 23404400 Free PMC article.
-
Origin and evolution of the sodium -pumping NADH: ubiquinone oxidoreductase.PLoS One. 2014 May 8;9(5):e96696. doi: 10.1371/journal.pone.0096696. eCollection 2014. PLoS One. 2014. PMID: 24809444 Free PMC article.
-
The conformational changes induced by ubiquinone binding in the Na+-pumping NADH:ubiquinone oxidoreductase (Na+-NQR) are kinetically controlled by conserved glycines 140 and 141 of the NqrB subunit.J Biol Chem. 2014 Aug 22;289(34):23723-33. doi: 10.1074/jbc.M114.574640. Epub 2014 Jul 8. J Biol Chem. 2014. PMID: 25006248 Free PMC article.
-
The Kinetic Reaction Mechanism of the Vibrio cholerae Sodium-dependent NADH Dehydrogenase.J Biol Chem. 2015 Aug 14;290(33):20009-21. doi: 10.1074/jbc.M115.658773. Epub 2015 May 23. J Biol Chem. 2015. PMID: 26004776 Free PMC article.
-
The TP0796 lipoprotein of Treponema pallidum is a bimetal-dependent FAD pyrophosphatase with a potential role in flavin homeostasis.J Biol Chem. 2013 Apr 19;288(16):11106-21. doi: 10.1074/jbc.M113.449975. Epub 2013 Feb 27. J Biol Chem. 2013. PMID: 23447540 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

