BCL-3 degradation involves its polyubiquitination through a FBW7-independent pathway and its binding to the proteasome subunit PSMB1
- PMID: 20558726
- PMCID: PMC2919145
- DOI: 10.1074/jbc.M110.112128
BCL-3 degradation involves its polyubiquitination through a FBW7-independent pathway and its binding to the proteasome subunit PSMB1
Abstract
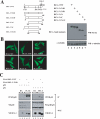
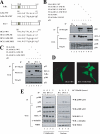
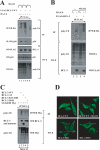
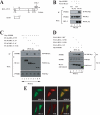
The oncogenic protein BCL-3 activates or represses gene transcription through binding with the NF-kappaB proteins p50 and p52 and is degraded through a phospho- and GSK3-dependent pathway. However, the mechanisms underlying its degradation remain poorly understood. Yeast two-hybrid analysis led to the identification of the proteasome subunit PSMB1 as a BCL-3-associated protein. The binding of BCL-3 to PSMB1 is required for its degradation through the proteasome. Indeed, PSMB1-depleted cells are defective in degrading polyubiquitinated BCL-3. The N-terminal part of BCL-3 includes lysines 13 and 26 required for the Lys(48)-linked polyubiquitination of BCL-3. Moreover, the E3 ligase FBW7, known to polyubiquitinate a variety of substrates phosphorylated by GSK3, is dispensable for BCL-3 degradation. Thus, our data defined a unique motif of BCL-3 that is needed for its recruitment to the proteasome and identified PSMB1 as a key protein required for the proteasome-mediated degradation of a nuclear and oncogenic IkappaB protein.
Figures
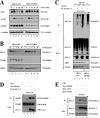
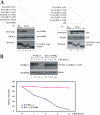

References
-
- Bours V., Franzoso G., Azarenko V., Park S., Kanno T., Brown K., Siebenlist U. (1993) Cell 72, 729–739 - PubMed
-
- Palmer S., Chen Y. H. (2008) Immunol. Res. 42, 210–218 - PubMed
-
- Kerr L. D., Duckett C. S., Wamsley P., Zhang Q., Chiao P., Nabel G., McKeithan T. W., Baeuerle P. A., Verma I. M. (1992) Genes Dev. 6(12A), 2352–2363 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

