Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes
- PMID: 20558737
- PMCID: PMC2924059
- DOI: 10.1074/jbc.M110.139931
Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes
Abstract
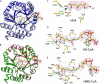
HMG-CoA lyase (HMGCL) is crucial to ketogenesis, and inherited human mutations are potentially lethal. Detailed understanding of the HMGCL reaction mechanism and the molecular basis for correlating human mutations with enzyme deficiency have been limited by the lack of structural information for enzyme liganded to an acyl-CoA substrate or inhibitor. Crystal structures of ternary complexes of WT HMGCL with the competitive inhibitor 3-hydroxyglutaryl-CoA and of the catalytically deficient HMGCL R41M mutant with substrate HMG-CoA have been determined to 2.4 and 2.2 A, respectively. Comparison of these beta/alpha-barrel structures with those of unliganded HMGCL and R41M reveals substantial differences for Mg(2+) coordination and positioning of the flexible loop containing the conserved HMGCL "signature" sequence. In the R41M-Mg(2+)-substrate ternary complex, loop residue Cys(266) (implicated in active-site function by mechanistic and mutagenesis observations) is more closely juxtaposed to the catalytic site than in the case of unliganded enzyme or the WT enzyme-Mg(2+)-3-hydroxyglutaryl-CoA inhibitor complex. In both ternary complexes, the S-stereoisomer of substrate or inhibitor is specifically bound, in accord with the observed Mg(2+) liganding of both C3 hydroxyl and C5 carboxyl oxygens. In addition to His(233) and His(235) imidazoles, other Mg(2+) ligands are the Asp(42) carboxyl oxygen and an ordered water molecule. This water, positioned between Asp(42) and the C3 hydroxyl of bound substrate/inhibitor, may function as a proton shuttle. The observed interaction of Arg(41) with the acyl-CoA C1 carbonyl oxygen explains the effects of Arg(41) mutation on reaction product enolization and explains why human Arg(41) mutations cause drastic enzyme deficiency.
Figures




Similar articles
-
Evaluation of 3-hydroxy-3-methylglutaryl-coenzyme A lyase arginine-41 as a catalytic residue: use of acetyldithio-coenzyme A to monitor product enolization.Biochemistry. 2004 May 11;43(18):5287-95. doi: 10.1021/bi0499765. Biochemistry. 2004. PMID: 15122894
-
Crystal structure of human 3-hydroxy-3-methylglutaryl-CoA Lyase: insights into catalysis and the molecular basis for hydroxymethylglutaric aciduria.J Biol Chem. 2006 Mar 17;281(11):7526-32. doi: 10.1074/jbc.M506880200. Epub 2005 Dec 5. J Biol Chem. 2006. PMID: 16330550
-
Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.J Biol Chem. 2006 Mar 17;281(11):7533-45. doi: 10.1074/jbc.M507996200. Epub 2005 Dec 5. J Biol Chem. 2006. PMID: 16330546
-
More Than One HMG-CoA Lyase: The Classical Mitochondrial Enzyme Plus the Peroxisomal and the Cytosolic Ones.Int J Mol Sci. 2019 Dec 4;20(24):6124. doi: 10.3390/ijms20246124. Int J Mol Sci. 2019. PMID: 31817290 Free PMC article. Review.
-
Molecular genetics of HMG-CoA lyase deficiency.Mol Genet Metab. 2007 Nov;92(3):198-209. doi: 10.1016/j.ymgme.2007.06.020. Epub 2007 Aug 9. Mol Genet Metab. 2007. PMID: 17692550 Review.
Cited by
-
Metabonomic analysis of potential biomarkers and drug targets involved in diabetic nephropathy mice.Sci Rep. 2015 Jul 7;5:11998. doi: 10.1038/srep11998. Sci Rep. 2015. PMID: 26149603 Free PMC article.
-
Characterization of a novel HMG-CoA lyase enzyme with a dual location in endoplasmic reticulum and cytosol.J Lipid Res. 2012 Oct;53(10):2046-2056. doi: 10.1194/jlr.M025700. Epub 2012 Jul 30. J Lipid Res. 2012. PMID: 22847177 Free PMC article.
-
miR-1202 regulates BPH-1 cell proliferation, apoptosis, and epithelial-to-mesenchymal transition through targeting HMGCL.Acta Biochim Biophys Sin (Shanghai). 2024 May 25;56(5):675-687. doi: 10.3724/abbs.2024001. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 38551020 Free PMC article.
-
Prevention of Dietary-Fat-Fueled Ketogenesis Attenuates BRAF V600E Tumor Growth.Cell Metab. 2017 Feb 7;25(2):358-373. doi: 10.1016/j.cmet.2016.12.010. Epub 2017 Jan 12. Cell Metab. 2017. PMID: 28089569 Free PMC article.
-
Influence of multiple cysteines on human 3-hydroxy-3-methylglutaryl-CoA lyase activity and formation of inter-subunit adducts.Arch Biochem Biophys. 2011 Jul;511(1-2):48-55. doi: 10.1016/j.abb.2011.04.004. Epub 2011 Apr 13. Arch Biochem Biophys. 2011. PMID: 21514269 Free PMC article.
References
-
- Stegink L. D., Coon M. J. (1968) J. Biol. Chem. 243, 5272–5279 - PubMed
-
- Robinson A. M., Williamson D. H. (1980) Physiol. Rev. 60, 143–187 - PubMed
-
- Wang S. P., Marth J. D., Oligny L. L., Vachon M., Robert M. F., Ashmarina L., Mitchell G. A. (1998) Hum. Mol. Genet. 7, 2057–2062 - PubMed
-
- Gibson K. M., Breuer J., Nyhan W. L. (1988) Eur. J. Pediatr. 148, 180–186 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

