Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis
- PMID: 20559436
- PMCID: PMC2885426
- DOI: 10.1371/journal.pone.0011113
Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis
Abstract
Background: Extracellular vesicles in yeast cells are involved in the molecular traffic across the cell wall. In yeast pathogens, these vesicles have been implicated in the transport of proteins, lipids, polysaccharide and pigments to the extracellular space. Cellular pathways required for the biogenesis of yeast extracellular vesicles are largely unknown.
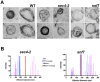
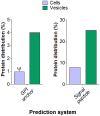
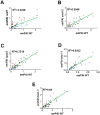
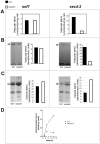
Methodology/principal findings: We characterized extracellular vesicle production in wild type (WT) and mutant strains of the model yeast Saccharomyces cerevisiae using transmission electron microscopy in combination with light scattering analysis, lipid extraction and proteomics. WT cells and mutants with defective expression of Sec4p, a secretory vesicle-associated Rab GTPase essential for Golgi-derived exocytosis, or Snf7p, which is involved in multivesicular body (MVB) formation, were analyzed in parallel. Bilayered vesicles with diameters at the 100-300 nm range were found in extracellular fractions from yeast cultures. Proteomic analysis of vesicular fractions from the cells aforementioned and additional mutants with defects in conventional secretion pathways (sec1-1, fusion of Golgi-derived exocytic vesicles with the plasma membrane; bos1-1, vesicle targeting to the Golgi complex) or MVB functionality (vps23, late endosomal trafficking) revealed a complex and interrelated protein collection. Semi-quantitative analysis of protein abundance revealed that mutations in both MVB- and Golgi-derived pathways affected the composition of yeast extracellular vesicles, but none abrogated vesicle production. Lipid analysis revealed that mutants with defects in Golgi-related components of the secretory pathway had slower vesicle release kinetics, as inferred from intracellular accumulation of sterols and reduced detection of these lipids in vesicle fractions in comparison with WT cells.
Conclusions/significance: Our results suggest that both conventional and unconventional pathways of secretion are required for biogenesis of extracellular vesicles, which demonstrate the complexity of this process in the biology of yeast cells.
Conflict of interest statement
Figures


Similar articles
-
Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle.Mol Biol Cell. 1997 Aug;8(8):1481-99. doi: 10.1091/mbc.8.8.1481. Mol Biol Cell. 1997. PMID: 9285820 Free PMC article.
-
In vitro reconstitution of Rab GTPase-dependent vesicle clustering by the yeast lethal giant larvae/tomosyn homolog, Sro7.J Biol Chem. 2015 Jan 2;290(1):612-24. doi: 10.1074/jbc.M114.595892. Epub 2014 Nov 17. J Biol Chem. 2015. PMID: 25404740 Free PMC article.
-
Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment.J Cell Biol. 1997 May 5;137(3):563-80. doi: 10.1083/jcb.137.3.563. J Cell Biol. 1997. PMID: 9151665 Free PMC article.
-
Functional morphology of the secretory pathway organelles in yeast.Microsc Res Tech. 2000 Dec 15;51(6):530-46. doi: 10.1002/1097-0029(20001215)51:6<530::AID-JEMT4>3.0.CO;2-Q. Microsc Res Tech. 2000. PMID: 11169856 Review.
-
Cytochemical images of secretion in Saccharomyces cerevisiae and animal cells are different.Acta Histochem. 1998 Nov;100(4):419-38. doi: 10.1016/S0065-1281(98)80039-1. Acta Histochem. 1998. PMID: 9842421 Review.
Cited by
-
Therapeutic applications of extracellular vesicles: clinical promise and open questions.Annu Rev Pharmacol Toxicol. 2015;55:439-464. doi: 10.1146/annurev-pharmtox-010814-124630. Epub 2014 Oct 3. Annu Rev Pharmacol Toxicol. 2015. PMID: 25292428 Free PMC article. Review.
-
Co-culturing experiments reveal the uptake of myo-inositol phosphate synthase (EC 5.5.1.4) in an inositol auxotroph of Saccharomyces cerevisiae.Microb Cell Fact. 2021 Jul 19;20(1):138. doi: 10.1186/s12934-021-01610-6. Microb Cell Fact. 2021. PMID: 34281557 Free PMC article.
-
Regulation of the fungal secretome.Curr Genet. 2016 Aug;62(3):533-45. doi: 10.1007/s00294-016-0578-2. Epub 2016 Feb 15. Curr Genet. 2016. PMID: 26879194 Review.
-
Updates in Paracoccidioides Biology and Genetic Advances in Fungus Manipulation.J Fungi (Basel). 2021 Feb 4;7(2):116. doi: 10.3390/jof7020116. J Fungi (Basel). 2021. PMID: 33557381 Free PMC article. Review.
-
Automated quantification of vacuole fusion and lipophagy in Saccharomyces cerevisiae from fluorescence and cryo-soft X-ray microscopy data using deep learning.Autophagy. 2024 Apr;20(4):902-922. doi: 10.1080/15548627.2023.2270378. Epub 2023 Oct 31. Autophagy. 2024. PMID: 37908116 Free PMC article.
References
-
- Glick BS, Malhotra V. The curious status of the Golgi apparatus. Cell. 1998;95:883–889. - PubMed
-
- Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155. - PubMed
-
- Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. - PubMed
-
- Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, et al. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. - PubMed
-
- Schekman R. Lasker Basic Medical Research Award. SEC mutants and the secretory apparatus. Nat Med. 2002;8:1055–1058. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AI033142/AI/NIAID NIH HHS/United States
- R01 AI033774/AI/NIAID NIH HHS/United States
- R01 AI056070/AI/NIAID NIH HHS/United States
- D43 TW007129/TW/FIC NIH HHS/United States
- R01 HL059842/HL/NHLBI NIH HHS/United States
- HL059842/HL/NHLBI NIH HHS/United States
- AI52733-06A1/AI/NIAID NIH HHS/United States
- R37 AI033142/AI/NIAID NIH HHS/United States
- G12 RR008124/RR/NCRR NIH HHS/United States
- #5G12RR008124-16A1/RR/NCRR NIH HHS/United States
- AI033142/AI/NIAID NIH HHS/United States
- AI033774/AI/NIAID NIH HHS/United States
- 5G12RR008124-16A1/RR/NCRR NIH HHS/United States
- R01 AI052733/AI/NIAID NIH HHS/United States
- AI052733/AI/NIAID NIH HHS/United States
- T32 GM007491/GM/NIGMS NIH HHS/United States
- 5G12RR008124-16A1S1/RR/NCRR NIH HHS/United States
- D43-TW007129/TW/FIC NIH HHS/United States
- AI056070-01A2/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

